This study examines the roles of microRNAs (miRNAs) in hepatocellular carcinoma (HCC), one of the most dangerous and common cancers globally. HCC has been widely studied, and its oncogenesis and progression is well understood. Although many therapeutic approaches have been developed, the prognosis for HCC, in terms of late diagnosis, resistance to chemotherapy, tumor recurrence, and metastasis, remains poor. Malignancy causes miRNAs to become deregulated, with widespread consequences in terms of cancer development and prognosis; miRNAs are therefore key therapeutic targets. This comprehensive review summarizes the roles of miRNAs in HCC, and examines their mechanisms of action. We believe that this review makes a significant contribution to the literature because we present an up-to-date and detailed overview of the involvement of miRNAs in HCC, and of their potential as therapeutic targets and biomarkers.
- microRNA
- hepatocellular carcinoma
- biomarker
1. Introduction
1.1. Hepatocellular Carcinoma
Liver cancer is the fourth leading cause of cancer deaths globally, after lung, colorectal, and stomach cancers [1]. Hepatocellular carcinoma (HCC) is the major subtype, accounting for more than 80% of primary liver cancers [2]. Multiple risk factors are associated with the occurrence of HCC, including chronic viral hepatitis B (HBV) or C (HCV) infections, which account for 80% of HCC cases worldwide [3]. Although alcoholic liver disease (ALD) is a common cause of HCC in the USA and Europe, the numbers of patients with nonalcoholic fatty liver disease (NAFLD) have recently increased in most developed countries, and NAFLD is now frequently listed as a cause of HCC [4][5]. Great therapeutic advances have been made in recent decades; however, the prognosis for HCC patients remains poor with a 5-year survival rate of 15–38% in the USA and Asia, attributed to late diagnosis, chemotherapy failure, and frequent recurrence [6][7][8]. Therefore, exploring the detailed mechanisms underlying HCC development and progression is an effective means of improving the outcomes of HCC patients.
1.2. MicroRNAs
Non-coding RNAs lack sequences that encode proteins or peptides; these are classified into small non-coding RNAs of about 20–30 bases and long non-coding RNAs of several hundred kbp. Small non-coding RNAs include microRNAs (miRNAs), small interfering RNA (siRNA), and PIWI-interacting RNA (piRNA). Among these, miRNAs are endogenous, small non-coding RNAs (approximately 19–25 nucleotides in length) that regulate gene expression through degradation of messenger RNAs (mRNAs) or inhibition of translation by binding to the 3′ untranslated region (UTR) of target mRNAs. miRNAs control cell proliferation, migration, invasion, and development in HCC by acting as tumor promoters or suppressors [9][10]. Each miRNA is thought to be capable of post-transcriptionally repressing hundreds of target genes; they are thus strong regulators of gene expression.
The biogenesis of miRNAs, and the mechanisms whereby they regulate translation by binding their target mRNAs, are illustrated in Figure 1. Primary miRNAs (pri-miRNA) are transcribed from DNA in the nucleus by RNA polymerase II and are approximately 3–4 kilobases long with a hairpin structure. They are processed by a nuclear RNase III enzyme (Drosha) and its partner protein DiGeorge syndrome critical region 8 (DGCR8), resulting in intermediate pre-miRNAs of approximately 70 nucleotides in length. Then, exportin-5 and its partner Ran-GTP bind to the pre-miRNA, and the complex is exported from the nucleus to the cytoplasm. The hairpin pre-miRNA is cleaved by ribonuclease III (Dicer) and the transactivation response element RNA-binding protein (TRBP), and is processed into a double-stranded mature miRNA of approximately 22 nucleotides in length. The double-stranded miRNA is then unwound, and one strand forms the RNA-induced silencing complex (RISC) with argonaute (Ago). The miRNA separates into mature, single-stranded miRNA, which is selected depending upon its stability, while less stable miRNA strands are degraded. Complementary base pairing between the seed region of the mature miRNA and the target mRNA results in the degradation of the mRNAs and translational repression [11][12].
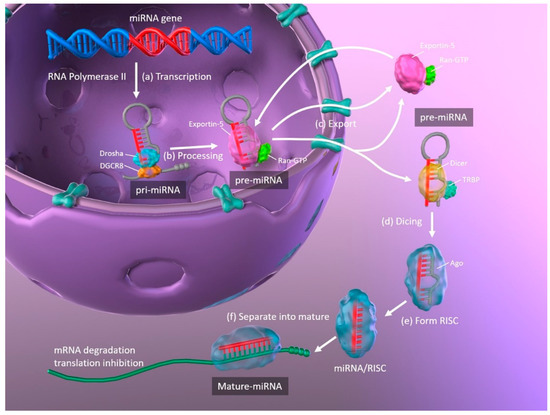
Figure 1. Schematic representation of miRNA biogenesis. (a) Synthesis of pri-miRNA transcripts from DNA. (b) pri-miRNA is processed by Drosha and DGCR8, resulting in a hairpin intermediate pre-miRNA. (c) Exportin-5 and Ran-GTP bind to pre-miRNA, and the complex is exported from the nucleus to the cytoplasm. (d) The hairpin pre-miRNA is cleaved by Dicer and TRBP. (e) The double-stranded miRNA is unwound and forms RISC with Ago. (f) The miRNA separates into mature, single-stranded miRNA. miRNA: microRNA; pri-miRNA: primary microRNA; DGCR8: DiGeorge syndrome critical region 8; TRBP: transactivation response element RNA-binding protein; RISC: RNA-induced silencing complex; Ago: argonaute.
miRNAs regulate approximately 30% of human genes via the pathway described above, many of which regulate various carcinogenic molecules and pathways or are located on unstable loci [13][14]. miRNAs have been shown to be dysregulated in most cancers. Oncogenic miRNAs (OncomiRs) and tumor-suppressive miRNAs are associated with carcinogenesis and malignant transformation contributing to or repressing the cancer phenotype. Overexpression of oncomiRs have been observed in various cancers [15]. Dysregulation of miRNAs in cancer has frequently been described in gastrointestinal [16][17], urological [18][19], gynecological [20], and lung cancers [21]. Increasing numbers of recent reports have described the utility of miRNAs as biomarkers or therapeutic targets in HCC [22][23].
2. The Role of miRNAs in Liver Regeneration
The liver has an exceptionally high regeneration capacity compared to other human organs. Therefore, in cases of liver damage, hepatocyte loss after hepatectomy, or secondary liver dysfunction, hepatocytes in the remaining liver tissue expand and proliferate to restore liver volume and function. Liver regeneration is a complex and regulated phenomenon in which multiple cell types that make up the liver interact via various signaling factors. Although the mechanisms of liver regeneration in the injured liver are clinically important, much remains unknown. Here, we summarized recent findings on the roles and importance of cell proliferation and apoptosis in liver regeneration.
2.1. Cell Proliferation in Hepatocytes
During regeneration and repair following liver injury, hepatocytes undergo division in three stages—initiation, growth, and termination. During initiation, hepatocytes are stimulated by cytokines such as interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) shift from the G0 cell-cycle phase to the G1 phase. In the subsequent growth phase, G1-phase hepatocytes proliferate by promoting the cell cycle in a cyclin-dependent manner, under stimulation by the hepatocyte growth factor (HGF). When liver volume and function return to normal after the growth phase, hepatocytes are stimulated by TGF-β and actin to terminate proliferation and return to the G0 phase [24]. Liver regeneration is precisely controlled by various molecular mechanisms. In particular, miRNAs have recently been documented to play key roles in the processes of cell proliferation [25][26][27][28][29].
The role of miRNAs and general miRNA changes during the initiation phase has long been known. One study revealed that approximately 40% of miRNAs had increased 3 h after partial hepatectomy, including those that targeted miRNA synthesis (Drosha, DGCR8, Dicer, and TRBP), whereas approximately 70% had declined at 24 h [25]. A transient increase in miRNAs acts as an initiation signal for cell proliferation, thus providing negative feedback that causes general miRNA changes after 24 h and promotes cell proliferation.
During the growth phase, the abundance of several miRNAs also declines; the levels of most miRNAs were lower within 3 d after partial hepatectomy, and gene expression related to cell cycle and proliferation were accordingly higher [26]. In contrast, several studies have reported increases in miRNA abundance during the growth phase, in which increased miR-21 plays an important role. miR-21 has been shown in vitro to control the transition from G1 to the S phase in primary hepatocyte proliferation and accelerate rapid S-phase entry by targeting a negative cell proliferation regulator, phosphatase, and tensin homolog deleted on the chromosome (PTEN) [27]. Further, an in vivo study demonstrated that miR-21 is a critical regulator of liver regeneration and its upregulation contributed to hepatocyte proliferation by targeting PTEN [28]. It was recently shown that the loss of Dicer1 inhibited liver regeneration by downregulating Dicer1-dependent miRNAs, including miR-21. Further, the introduction of miR-21 restored liver regeneration by inhibiting PTEN and Ras homolog family member B (RhoB). RhoB facilitates activation of the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) axis [29].
Many miRNAs suppress the proliferation of hepatocytes after the initiation phase and induce liver regeneration until the termination phase is reached. miR-23b, which promotes cell proliferation by small mother against decapentaplegic targeting 3 (SMAD3) and inhibiting the TGF-β pathway, was highly expressed up to 24 h after partial hepatectomy; however, its expression subsequently declined consistently from 3 to 7 days [30]. In contrast, miR-34a, which acts as a growth inhibitor, was consistently overexpressed up to 24 h after partial hepatectomy [31]. Blocking or stimulating miRNA pathways during liver regeneration may provide novel therapeutic strategies for managing liver regeneration [32].
2.2. Apoptosis in Hepatocytes
Apoptosis refers to highly regulated cell death; it occurs not only in morphogenesis during development, but also serves to control cell numbers and to remove damaged cells in liver steatosis, inflammation, and fibrosis. It, therefore, plays an important role in hepatocarcinogenesis. Several miRNAs regulate programmed cell death via intrinsic (BCL2 and MCL1) and extrinsic (TNF-related apoptosis-inducing ligand; TRAIL and Fas) regulatory pathways and the p53-mediated and endoplasmic reticulum stress-induced apoptosis pathway [33]. The following section describes the primary roles of miRNAs in regulating the key molecules involved in the hepatocyte apoptotic pathway.
miR-15b and miR-16 are TNF-related apoptosis regulators via the antiapoptotic protein BCL2. In severe liver inflammation, miR-15b and miR-16 were upregulated in liver tissues and regulated BCL2 at the protein level; moreover, inhibition of these miRNAs reduced TNF-mediated apoptosis in the liver [34]. Recent evidence indicates that miR-15b can inhibit cell proliferation and downregulate BCL2 mRNA and protein expression in hepatocytes [35].
Abnormal expression of miR-125b is common in various cancers; miR-125b plays dual roles to induce or inhibit apoptosis, depending on the cellular state. Similarly, miR-125b downregulation is frequently observed in HCC development; low expression of miR-125b in HCC tissues correlates with a higher rate of apoptosis [36]. miR-125b is thought to promote apoptosis by directly targeting BCL family members such as MCL1, BCLw, and IL-6R [36]. Therefore, miR125b downregulation may promote malignant transformation and tumor development.
miR-221 and its paralog miRNA-222 have been shown in several studies to aggravate hepatocarcinogenesis by targeting apoptosis-related factors, such as p53, p53 upregulated modulator of apoptosis (PUMA), NF-κB, and signal transducer and activator of transcription 3 (STAT3) [37][38]. During hepatocellular carcinogenesis in vitro and in vivo, miR-221/222 induced TRAIL resistance by regulating tumor suppressors such as PTEN and tissue inhibitor of metalloproteinase 3 (TIMP3), and enhanced cell migration by activating the AKT pathway and metallopeptidase activity [39]. In mice, overexpression of miR-221 led to antiapoptotic effects and delayed liver failure progression by regulating the expression of PUMA in the liver [40].
3. Dysregulation of miRNAs in HCC
Each miRNA targets multiple mRNAs and regulates the expression of multiple genes in a complex manner; these dysregulations are observed in various cancer types [16][17][18][19][20][21][41]. While some miRNAs are downregulated in HCC, acting as tumor suppressors, upregulated miRNAs also occur in HCC, functioning as oncomiRs. Regions encoding miRNAs involved in dysfunction can harbor genetic alterations such as mutations, amplifications, deletions, or fusions. Transcription of some miRNAs is suppressed by carcinogenesis transcription factors such as Myc, while others are epigenetically regulated by DNA methylation and histone modifications. Additionally, suppression of miRNA processing machinery genes, including Drosha, DGCR8, Dicer, TRBP, and Ago2 has been shown to reduce mature miRNA synthesis, leading to hepatocarcinogenesis and HCC development [41].
In HCC, miRNA dysregulation is involved in all clinical stages, and is evident even in early stages; therefore, miRNA profiles may potentially be used to discriminate HCC patients from healthy controls and those with other liver diseases [42]. Various HCC-specific miRNA signatures have been recently identified. In a previous study, we reported that miRNA profiles differed between hepatocytes and HCC cell lines [43], suggesting that miRNA profiles have significant value as biomarkers [44][45]. Insights into the roles of miRNAs in HCC development and progression have made miRNAs attractive biomarkers and therapeutic targets for HCC.
4. Conclusions
The biological significance and utility of miRNAs in liver disease, especially in HCC, is a rapidly growing field. Accumulating evidence has demonstrated that many miRNAs play regulatory roles in many biological processes associated with HCC, including oncogenesis, tumor development, cell proliferation, and apoptosis. In this article, we reviewed the molecular and functional roles of miRNAs in the development and progression of HCC. Many studies have investigated aberrant miRNA processing and miRNA expression profiles in HCC, including those of circulating miRNAs, which have contributed to the discovery and clinical adaptation of miRNAs as potential biomarkers for diagnosis (particularly early diagnosis) and as prognostic markers for HCC. Furthermore, investigating their molecular and functional roles is likely to be useful for developing therapeutic targets and understanding resistance to conventional therapies. Although miRNAs do not have a direct antitumor effect on HCC, miRNA-based therapy offers a promising perspective compared to classical gene therapy such as induction of a single gene because miRNA exerts antitumor effects by regulating the expression of multiple genes involved in hepatocarcinogenesis. In addition, miRNAs are generally not immunogenic because they do not encode proteins. A combination of conventional and miRNA-based therapy for HCC may be preferable by offering improved gene transfer efficiency and transgene expression.
Although miRNAs have demonstrated potential as biological targets for HCC treatment in preclinical studies, miRNA-based therapy is not yet suitable for clinical practice. Some important problems need to be resolved before clinical application. The first is that due to tumor cell heterogeneity, miRNAs exert different regulatory effects in different types of tumors, and may even show opposite results in different studies. The second is that in vivo research with miRNAs is still relatively scarce; miRNA regulation in vivo may not always be observed due to the complexity of the in-vivo environment. Therefore, extensive in vivo confirmation of miRNA roles in the progression of HCC and their therapeutic effects is required. Finally, because miRNAs are large molecules, further research is needed to effectively administer and deliver miRNAs into tumor cells in the body. Ongoing global miRNA research will be useful for understanding the current state of clinical miRNA application, and for elucidating the biological characteristics underlying diagnosis, treatment, pathology, and prognosis prediction in HCC.
This entry is adapted from the peer-reviewed paper 10.3390/ijms21218362
References
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017, 3, 5245–48.
- El-Serag, H.B.; Rudolph, K.L.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576.
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604.
- Park, J.W.; Chen, M.; Colombo, M.; Roberts, L.R.; Schwartz, M.; Chen, P.J.; Kudo, M.; Johnson, P.; Wagner, S.; Orsini, L.S.; et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015, 35, 2155–2166.
- Younossi, Z.M.; Blissett, D.; Blissett, R.; Henry, L.; Stepanova, M.; Younossi, Y.; Racila, A.; Hunt, S.; Beckerman, R.R. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 2016, 64, 1577–1586.
- Altekruse, S.F.; Henley, S.J.; Cucinelli, J.E.; McGlynn, K.A.A. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am. J. Gastroenterol. 2014, 109, 542–553.
- Zhang, G.; Li, R.; Deng, Y.; Zhao, L.L. Conditional survival of patients with hepatocellular carcinoma: Results from the Surveillance, Epidemiology, and End Results registry. Expert Rev. Gastroenterol. Hepatol. 2018, 12, 515–523.
- Xu, L.; Kim, Y.; Spolverato, G.; Gani, F.; Pawlik, T.M.M. Racial disparities in treatment and survival of patients with hepatocellular carcinoma in the United States. Hepatobiliary Surg. Nutr. 2016, 5, 43–52.
- Esquela-Kerscher, A.; Slack, F.J.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 2596–9.
- Bartel, D.P.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297.
- Kim, V.N.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385.
- Borchert, G.M.; Lanier, W.; Davidson, B.L.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101.
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587.
- Si, W.; Shen, J.; Zheng, H.; Fan, W.W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 2019, 11, 25.
- Cheng, C.J.; Slack, F.J.J. The duality of oncomiR addiction in the maintenance and treatment of cancer. Cancer J. 2012, 18, 232–237.
- Nedaeinia, R.; Manian, M.; Jazayeri, M.H.; Ranjbar, M.; Salehi, R.; Sharifi, M.; Mohaghegh, F.; Goli, M.; Jahednia, S.H.; Avan, A.; et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2017, 24, 48–56.
- Zheng, Q.; Chen, C.; Guan, H.; Kang, W.; Yu, C.C. Prognostic role of microRNAs in human gastrointestinal cancer: A systematic review and meta-analysis. Oncotarget 2017, 8, 46611–46623.
- Santoni, G.; Morelli, M.B.; Amantini, C.; Battelli, N.N. Urinary markers in bladder cancer: An update. Front. Oncol. 2018, 8, 362.
- Wang, J.; Ni, J.; Beretov, J.; Thompson, J.; Graham, P.; Li, Y.Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol. 2020, 145, 102860.
- Miao, J.; Regenstein, J.M.; Xu, D.; Zhou, D.; Li, H.; Zhang, H.; Li, C.; Qiu, J.; Chen, X.X. The roles of microRNA in human cervical cancer. Arch. Biochem. Biophys. 2020, 690, 108480.
- Wadowska, K.; Bil-Lula, I.; Trembecki, L.; Sliwinska-Mosson, M.M. Genetic markers in lung cancer diagnosis: A review. Int. J. Mol. Sci. 2020, 21, 4569.
- Xu, J.; An, P.; Winkler, C.A.; Yu, Y.Y. Dysregulated microRNAs in hepatitis B virus-related hepatocellular carcinoma: Potential as biomarkers and therapeutic targets. Front. Oncol. 2020, 10, 1271.
- Lim, L.J.; Wong, S.Y.S.; Huang, F.; Lim, S.; Chong, S.S.; Ooi, L.L.; Kon, O.L.; Lee, C.G.G. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. 2019, 79, 5131–5139.
- Fausto, N.; Campbell, J.S.; Riehle, K.J.J. Liver regeneration. Hepatology 2006, 43, (Suppl. 1), S45–S53.
- Shu, J.; Kren, B.T.; Xia, Z.; Wong, P.Y.; Li, L.; Hanse, E.A.; Min, M.X.; Li, B.; Albrecht, J.H.; Zeng, Y.; et al. Genomewide microRNA down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology 2011, 54, 609–619.
- Chen, X.; Murad, M.; Cui, Y.Y.; Yao, L.J.; Venugopal, S.K.; Dawson, K.; Wu, J.J. miRNA regulation of liver growth after 50% partial hepatectomy and small size grafts in rats. Transplantation 2011, 91, 293–399.
- Yan-nan, B.; Zhao-yan, Y.; Li-xi, L.; Jiang, Y.; Qing-jie, X.; Yong, Z.Z. MicroRNA-21 accelerates hepatocyte proliferation in vitro via PI3K/Akt signaling by targeting PTEN. Biochem. Biophys. Res. Commun. 2014, 443, 802–807.
- Chen, X.; Song, M.; Chen, W.; Dimitrova-Shumkovska, J.; Zhao, Y.; Cao, Y.; Song, Y.; Yang, W.; Wang, F.; Xiang, Y.; et al. MicroRNA-21 contributes to liver regeneration by targeting PTEN. Med. Sci. Monit. 2016, 22, 83–91.
- Lv, T.; Kong, L.; Jiang, L.; Wu, H.; Wen, T.; Shi, Y.; Yang, J.J. Dicer1 facilitates liver regeneration in a manner dependent on the inhibitory effect of miR-21 on Pten and Rhob expression. Life Sci. 2019, 232, 116656.
- Yuan, B.; Dong, R.; Shi, D.; Zhou, Y.; Zhao, Y.; Miao, M.; Jiao, B.B. Down-regulation of miR-23b may contribute to activation of the TGF-beta1/Smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett. 2011, 585, 9273–4.
- Chen, H.; Sun, Y.; Dong, R.; Yang, S.; Pan, C.; Xiang, D.; Miao, M.; Jiao, B.B. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS ONE 2011, 6, e20238.
- Yi, P.S.; Zhang, M.; Xu, M.Q.Q. Role of microRNA in liver regeneration. Hepatobiliary Pancreat. Dis. Int. 2016, 15, 141–146.
- Shirjang, S.; Mansoori, B.; Asghari, S.; Duijf, P.H.G.; Mohammadi, A.; Gjerstorff, M.; Baradaran, B.B. MicroRNAs in cancer cell death pathways: Apoptosis and necroptosis. Free Radic. Biol. Med. 2019, 139, 1–15.
- An, F.; Gong, B.; Wang, H.; Yu, D.; Zhao, G.; Lin, L.; Tang, W.; Yu, H.; Bao, S.; Xie, Q.Q. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis 2012, 17, 702–716.
- Zhang, Y.; Huang, F.; Wang, J.; Peng, L.; Luo, H.H. MiR-15b mediates liver cancer cells proliferation through targeting BCL-2. Int. J. Clin. Exp. Pathol. 2015, 8, 15677–15683.
- Gong, J.; Zhang, J.P.; Li, B.; Zeng, C.; You, K.; Chen, M.X.; Yuan, Y.; Zhuang, S.M.M. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013, 32, 3071–3079.
- Santhekadur, P.K.; Das, S.K.; Gredler, R.; Chen, D.; Srivastava, J.; Robertson, C.; Baldwin, A.S.S., Jr.; Fisher, P.B.; Sarkar, D.D. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J. Biol. Chem. 2012, 287, 13952–13958.
- Li, Y.; Di, C.; Li, W.; Cai, W.; Tan, X.; Xu, L.; Yang, L.; Lou, G.; Yan, Y.Y. Oncomirs miRNA-221/222 and tumor suppressors miRNA-199a/195 are crucial miRNAs in liver cancer: A systematic analysis. Dig. Dis. Sci. 2016, 61, 2315–2327.
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16, 498–509.
- Sharma, A.D.; Narain, N.; Handel, E.M.; Iken, M.; Singhal, N.; Cathomen, T.; Manns, M.P.; Scholer, H.R.; Ott, M.; Cantz, T.T. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology 2011, 53, 1651–1661.
- Khan, S.; Ayub, H.; Khan, T.; Wahid, F.F. MicroRNA biogenesis, gene silencing mechanisms and role in breast, ovarian and prostate cancer. Biochimie 2019, 167, 12–24.
- Borel, F.; Konstantinova, P.; Jansen, P.L.L. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J. Hepatol. 2012, 56, 1371–1383.
- Morishita, A.; Iwama, H.; Fujihara, S.; Sakamoto, T.; Fujita, K.; Tani, J.; Miyoshi, H.; Yoneyama, H.; Himoto, T.; Masaki, T.T. MicroRNA profiles in various hepatocellular carcinoma cell lines. Oncol. Lett. 2016, 12, 1687–1692.
- Morishita, A.; Masaki, T.T. MicroRNAs as possible biomarkers for hepatocellular carcinoma. Hepatol. Res. 2018, 48, 499–501.
- Oura, K.; Fujita, K.; Morishita, A.; Iwama, H.; Nakahara, M.; Tadokoro, T.; Sakamoto, T.; Nomura, T.; Yoneyama, H.; Mimura, S.; et al. Serum microRNA-125a-5p as a potential biomarker of HCV-associated hepatocellular carcinoma. Oncol. Lett. 2019, 18, 8828–90.
