Helicobacter pylori is a gastric pathogen that infects nearly half of the global population and is recognized as a group 1 carcinogen by the Word Health Organization. The global rise in antibiotic resistance has increased clinical challenges in treating H. pylori infections. Biofilm growth has been proposed to contribute to H. pylori’s chronic colonization of the host stomach, treatment failures, and the eventual development of gastric diseases. Several components of H. pylori have been identified to promote biofilm growth, and several of these may also facilitate antibiotic tolerance, including the extracellular matrix, outer membrane proteins, shifted morphology, modulated metabolism, efflux pumps, and virulence factors.
- Helicobacter pylori
- biofilms
- planktonic
- antibiotic resistance
- extra polymeric substance
1. Introduction
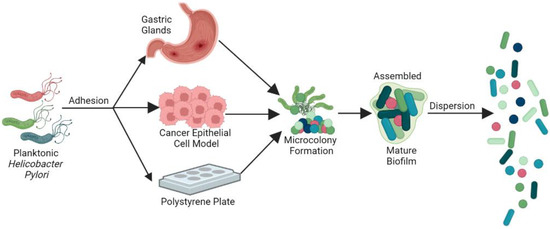
2. General Features of H. pylori Biofilms
3. H. pylori Clinical Treatment Strategies Become Less Efficient, Highlighting the Requirement of Alternative Strategies
4. Regulation in H. pylori Biofilm
This entry is adapted from the peer-reviewed paper 10.3390/antibiotics12081260
References
- Dubois, A.; Fiala, N.; Heman-Ackah, L.M.; Drazek, E.; Tarnawski, A.; Fishbein, W.N.; Perez-Perez, G.I.; Blaser, M.J. Natural gastric infection with Helicobacter pylori in monkeys: A model for spiral bacteria infection in humans. Gastroenterology 1994, 106, 1405–1417.
- Hooi, J.K.Y.; Lai, W.Y.; Ng, W.K.; Suen, M.M.Y.; Underwood, F.E.; Tanyingoh, D.; Malfertheiner, P.; Graham, D.Y.; Wong, V.W.S.; Wu, J.C.Y.; et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology 2017, 153, 420–429.
- Malfertheiner, P.; Camargo, M.C.; El-Omar, E.; Liou, J.M.; Peek, R.; Schulz, C.; Smith, S.I.; Suerbaum, S. Helicobacter pylori infection. Nat. Rev. Dis. Primers 2023, 9, 19.
- Labenz, J.; Borsch, G. Evidence for the essential role of Helicobacter pylori in gastric ulcer disease. Gut 1994, 35, 19–22.
- Blaser, M.J. Helicobacter pylori Phenotypes Associated with Peptic Ulceration. Scand. J. Gastroenterol. Suppl. 1994, 205, 1–5.
- Pereira, M.-I. Role of Helicobacter pylori in gastric mucosa-associated lymphoid tissue lymphomas. World J. Gastroenterol. 2014, 20, 684–698.
- Asaka, M.; Kimura, T.; Kato, M.; Kudo, M.; Miki, K.; Ogoshi, K.; Kato, T.; Tatsuta, M.; Graham, D.Y. Possible role of Helicobacter pylori infection in early gastric cancer development. Cancer 1994, 73, 2691–2694.
- van Zanten, S.J.V.; Sherman, P.M. Helicobacter pylori infection as a cause of gastritis, duodenal ulcer, gastric cancer and nonulcer dyspepsia: A systematic overview. CMAJ 1994, 150, 177–185.
- Isaacson, P.G. Gastric Lymphoma and Helicobacter pylori. N. Engl. J. Med. 1994, 330, 1310–1311.
- Ma, J.; Yu, M.; Shao, Q.; Yu, X.; Zhang, C.; Zhao, J.; Yuan, L.; Qi, Y.; Hu, R.; Wei, P.; et al. Both family-based Helicobacter pylori infection control and management strategy and screen-and-treat strategy are cost-effective for gastric cancer prevention. Helicobacter 2022, 27, e12911.
- Liu, Y.; Wang, S.; Yang, F.; Chi, W.; Ding, L.; Liu, T.; Zhu, F.; Ji, D.; Zhou, J.; Fang, Y.; et al. Antimicrobial resistance patterns and genetic elements associated with the antibiotic resistance of Helicobacter pylori strains from Shanghai. Gut Pathog. 2022, 14, 14.
- Karbalaei, M.; Keikha, M.; Abadi, A.T.B. Prevalence of Primary Multidrug-resistant Helicobac-ter pylori in Children: A Systematic Review and Meta-analysis. Arch. Med. Res. 2022, 53, 634–640.
- Brown, H.; Cantrell, S.; Tang, H.; Epplein, M.; Garman, K.S. Racial Differences in Helicobacter pylori Prevalence in the US: A Systematic Review. Gastro Hep Adv. 2022, 1, 857–868.
- Cellini, L.; Grande, R.; Di Campli, E.; Traini, T.; Di Giulio, M.; Lannutti, S.N.; Lattanzio, R. Dynamic colonization of Helicobacter pylori in human gastric mucosa. Scand. J. Gastroenterol. 2008, 43, 178–185.
- Mackay, W.; Gribbon, L.; Barer, M.; Reid, D. Biofilms in drinking water systems: A possible reservoir for Helicobacter pylori. J. Appl. Microbiol. 1998, 85, 52S–59S.
- Stark, R.M.; Gerwig, G.J.; Pitman, R.S.; Potts, L.F.; Williams, N.A.; Greenman, J.; Weinzweig, I.P.; Hirst, T.R.; Millar, M.R. Biofilm for-mation by Helicobacter pylori. Lett. Appl. Microbiol. 1999, 28, 121–126.
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter pylori biofilms in human gas-tric mucosa. J. Gastrointest. Surg. 2006, 10, 712–717.
- Coticchia, J.M.; Sugawa, C.; Tran, V.R.; Gurrola, J.; Kowalski, E.; Carron, M.A. Presence and Density of Helicobacter pylori Biofilms in Human Gastric Mucosa in Patients with Peptic Ulcer Disease. J. Gastrointest. Surg. 2006, 10, 883–889.
- Windham, I.H.; Servetas, S.L.; Whitmire, J.M.; Pletzer, D.; Hancock, R.E.; Merrell, D.S. Helicobacter pylori bio-film formation is differentially affected by common culture conditions, and proteins play a central role in the biofilm matrix. Appl. Environ. Microbiol. 2018, 84, e00391-18.
- Grande, R.; Di Giulio, M.; Bessa, L.J.; Di Campli, E.; Baffoni, M.; Guarnieri, S.; Cellini, L. Extracellular DNA in Hel-icobacter pylori biofilm: A backstairs rumour. J. Appl. Microbiol. 2011, 110, 490–498.
- Li, P.; Chen, X.; Shen, Y.; Li, H.; Zou, Y.; Yuan, G.; Hu, P.; Hu, H. Mucus penetration enhanced lipid polymer nanopar-ticles improve the eradication rate of Helicobacter pylori biofilm. J. Control. Release 2019, 300, 52–63.
- Cole, S.P.; Harwood, J.; Lee, R.; She, R.; Guiney, D.G. Characterization of monospecies biofilm formation by Heli-cobacter pylori. J. Bacteriol. 2004, 186, 3124–3132.
- Hathroubi, S.; Hu, S.; Ottemann, K.M. Genetic requirements and transcriptomics of Helicobacter pylori biofilm formation on abiotic and biotic surfaces. npj Biofilms Microbiomes 2020, 6, 56.
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Genet. 2022, 20, 608–620.
- Hathroubi, S.; Zerebinski, J.; Ottemann, K.M. Helicobacter pylori Biofilm Involves a Multigene Stress-Biased Response, Including a Structural Role for Flagella. mBio 2018, 9, e01973-18.
- Williams, J.C.; McInnis, K.A.; Testerman, T.L. Adherence of Helicobacter pylori to Abiotic Surfaces Is Influenced by Serum. Appl. Environ. Microbiol. 2008, 74, 1255–1258.
- Tan, S.; Tompkins, L.S.; Amieva, M.R. Helicobacter pylori Usurps Cell Polarity to Turn the Cell Surface into a Replicative Niche. PLOS Pathog. 2009, 5, e1000407.
- Anderson, J.K.; Huang, J.Y.; Wreden, C.; Sweeney, E.G.; Goers, J.; Remington, S.J.; Guillemin, K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. mBio 2015, 6, e00379-15.
- Sigal, M.; Rothenberg, M.E.; Logan, C.Y.; Lee, J.Y.; Honaker, R.W.; Cooper, R.L.; Passarelli, B.; Camorlinga, M.; Bouley, D.M.; Alvarez, G.; et al. Helicobacter pylori Activates and Expands Lgr5+ Stem Cells Through Direct Colonization of the Gastric Glands. Gastroenterology 2015, 148, 1392–1404.e21.
- Cellini, L.; Grande, R.; Di Campli, E.; Di Bartolomeo, S.; Di Giulio, M.; Traini, T.; Trubiani, O. Characterization of an Helicobacter pylori environmental strain. J. Appl. Microbiol. 2008, 105, 761–769.
- Bugli, F.; Palmieri, V.; Torelli, R.; Papi, M.; De Spirito, M.; Cacaci, M.; Galgano, S.; Masucci, L.; Paroni Sterbini, F.; Vella, A.; et al. In vitro effect of clarithromycin and alginate lyase against Helicobacter pylori biofilm. Biotechnol. Prog. 2016, 32, 1584–1591.
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.-M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762.
- Liu, W.Z.; Xie, Y.; Lu, H.; Cheng, H.; Zeng, Z.R.; Zhou, L.Y.; Chen, Y.; Bin Wang, J.; Du, Y.Q.; Lu, N.H. Fifth Chinese National Consensus Report on the management of Helicobacter pylori infection. Helicobacter 2018, 23, e12475.
- Pan, J.; Shi, Z.; Lin, D.; Yang, N.; Meng, F.; Lin, L.; Jin, Z.; Zhou, Q.; Wu, J.; Zhang, J.; et al. Is tailored therapy based on antibiotic susceptibility effective ? a multicenter, open-label, randomized trial. Front. Med. 2020, 14, 43–50.
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management of Helicobacter pylori infection—The Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30.
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17.
- Miftahussurur, M.; Syam, A.F.; Nusi, I.A.; Makmun, D.; Waskito, L.A.; Zein, L.H.; Akil, F.; Uwan, W.B.; Simanjuntak, D.; Wibawa, I.D.N.; et al. Surveillance of Helicobacter pylori Antibiotic Susceptibility in Indonesia: Different Resistance Types among Regions and with Novel Genetic Mutations. PLoS ONE 2016, 11, e0166199.
- Fauzia, K.A.; Miftahussurur, M.; Syam, A.F.; Waskito, L.A.; Doohan, D.; Rezkitha, Y.A.A.; Matsumoto, T.; Tuan, V.P.; Akada, J.; Yonezawa, H.; et al. Biofilm Formation and Antibiotic Resistance Phenotype of Helicobacter pylori Clinical Isolates. Toxins 2020, 12, 473.
- Fallone, C.A.; Chiba, N.; van Zanten, S.V.; Fischbach, L.; Gisbert, J.P.; Hunt, R.H.; Jones, N.L.; Render, C.; Leontiadis, G.I.; Moayyedi, P.; et al. The Toronto Consensus for the Treatment of Helicobacter pylori Infection in Adults. Gastroenterology 2016, 151, 51–69.e14.
- De Palma, G.Z.; Mendiondo, N.; Wonaga, A.; Viola, L.; Ibarra, D.; Campitelli, E.; Salim, N.; Corti, R.; Goldman, C.; Catalano, M. Occurrence of mutations in the antimicrobial target genes related to levofloxacin, clarithromycin, and amoxicillin re-sistance in Helicobacter pylori isolates from Buenos Aires city. Microb. Drug Resist. 2017, 23, 351–358.
- Li, H.; Shen, Y.; Song, X.; Tang, X.; Hu, R.; Marshall, B.J.; Tang, H.; Benghezal, M. Need for standardization and harmonization of Helicobacter pylori antimicrobial susceptibility testing. Helicobacter 2022, 27, e12873.
- Mascellino, M.T.; Oliva, A.; De Angelis, M.; Pontone, S.; Porowska, B. Helicobacter pylori infection: Antibiotic resistance and eradi-cation rate in patients with gastritis showing previous treatment failures. New Microbiol. 2018, 41, 306–309.
- Redondo, J.J.; Keller, P.M.; Zbinden, R.; Wagner, K. A novel RT-PCR for the detection of Heli-cobacter pylori and identification of clarithromycin resistance mediated by mutations in the 23S rRNA gene. Diagn. Microbiol. Infect. Dis. 2018, 90, 1–6.
- Lee, S.M.; Kim, N.; Kwon, Y.H.; Nam, R.H.; Kim, J.M.; Park, J.Y.; Lee, Y.S.; Lee, D.H. rdxA, frxA, and efflux pump in metronidazole-resistant Helicobacter pylori: Their relation to clinical out-comes. J. Gastroenterol. Hepatol. 2018, 33, 681–688.
- Miftahussurur, M.; Shrestha, P.K.; Subsomwong, P.; Sharma, R.P.; Yamaoka, Y. Emerging Helicobacter pylori levofloxacin resistance and novel genetic mutation in Nepal. BMC Microbiol. 2016, 16, 256.
- Hanafi, A.; Lee, W.C.; Loke, M.F.; Teh, X.; Shaari, A.; Dinarvand, M.; Lehours, P.; Mégraud, F.; Leow, A.H.R.; Vadivelu, J.; et al. Molecular and Proteomic Analysis of Levofloxacin and Metronidazole Resistant Helicobacter pylori. Front. Microbiol. 2016, 7, 2015.
- Attaran, B.; Falsafi, T. Identification of factors associated with biofilm formation ability in the clinical iso-lates of Helicobacter pylori. Iran. J. Biotechnol. 2017, 15, 58.
- Attaran, B.; Falsafi, T.; Ghorbanmehr, N. Effect of biofilm formation by clinical isolates of Helicobacter pylori on the efflux-mediated resistance to commonly used antibiotics. World J. Gastroenterol. 2017, 23, 1163.
- Ge, X.; Cai, Y.; Chen, Z.; Gao, S.; Geng, X.; Li, Y.; Jia, J.; Sun, Y. Bifunctional Enzyme SpoT Is Involved in Biofilm Formation of Helicobacter pylori with Multidrug Resistance by Upregulating Efflux Pump Hp1174 (gluP). Antimicrob. Agents Chemother. 2018, 62, e00957-18.
- Servetas, S.L.; Kim, A.; Su, H.; Cha, J.; Merrell, D.S. Comparative analysis of the Hom family of outer membrane proteins in isolates from two geographically distinct regions: The United States and South Korea. Helicobacter 2018, 23, e12461.
- Servetas, S.L.; Doster, R.S.; Kim, A.; Windham, I.H.; Cha, J.H.; Gaddy, J.A.; Merrell, D.S. ArsRS-dependent regulation of homB contributes to Helicobacter pylori biofilm formation. Front. Microbiol. 2018, 2, 1497.
- De la Cruz, M.A.; Ares, M.; Von Bargen, K.; Panunzi, L.G.; Martínez-Cruz, J.; Valdez-Salazar, H.-A.; Jiménez-Galicia, C.; Torres, J. Gene Expression Profiling of Transcription Factors of Helicobacter pylori under Different Environmental Conditions. Front. Microbiol. 2017, 8, 615.
