Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
SRC homology 3 (SH3) domains are fundamental modules that enable the assembly of protein complexes through physical interactions with a pool of proline-rich/noncanonical motifs from partner proteins. They are widely studied modular building blocks across all five kingdoms of life and viruses, mediating various biological processes. The SH3 domains are also implicated in the development of human diseases, such as cancer, leukemia, osteoporosis, Alzheimer’s disease, and various infections.
- proline-rich motifs (PRM)
- protein interaction
- SH3 domain
- SH3 domain-containing proteins
1. General Introduction
The SRC homology 3 (SH3) domain was first described in 1988 as a region of approximately 60 amino acids found in different intracellular signaling proteins, such as SRC and PLC [1][2]. SH3 domains are arranged as small protein modules in a compact β-barrel fold made of five β-strands connected by RT, n-SRC, distal loops, and a 310-helix (Figure 1) [3]. Thousands of SH3 domains present in eukaryotes, prokaryotes, and viruses have been investigated and characterized as modules mediating the protein–protein interaction/association [4][5]. SH3 domain-mediated protein–protein interactions have significant diversification as the binding partners regulate almost all essential cellular functions, including cell survival, proliferation, differentiation, migration, and polarity. Moreover, findings underscore the significance of SH3 domains in shaping protein–protein interaction, their potential influence on protein folding and positioning, their impact on cellular phenotypes, and the essential role they play in protein function [6]. Mutations and malfunctions of the SH3 domain can lead to significant neurological defects, cancer, and infectious diseases [7][8][9].
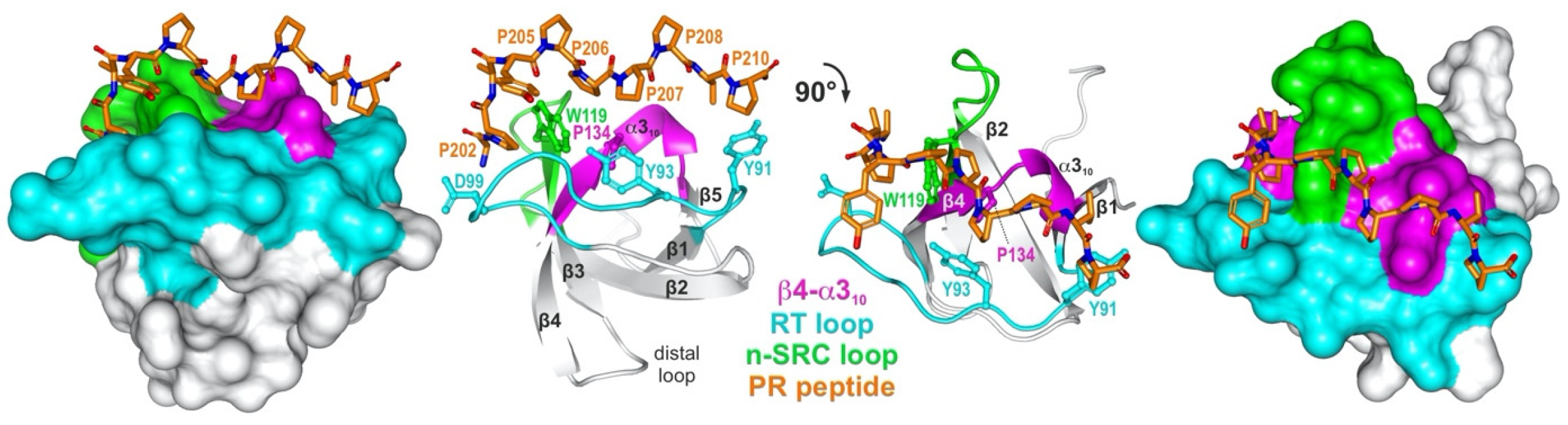
Figure 1. A representative structure of an SH3 domain PRM complex. A detailed view into the structure (PDB code: 1FYN) of the SH3 domain of FYN tyrosine kinase (left: surface representation; right: ribbon representation; UniProt ID: P06241) in complex with 3BP-2 PR peptide (PAYPPPPVP; orange; UniProt ID: P78314) which shows the characteristic arrangement of beta strands and the PRM-interacting variable loops, referred to as β4-α310 (magenta), RT (cyan), and hydrophobic patch (W1190) flanked by n-SRC loop (green). Conserved residues that are crucial for the interaction are Y91, Y93, D99, W119, and P134. FYN SH3 shows the typical topology of two perpendicular three-stranded β-sheets and a single turn of α310.
SH3 domain-containing proteins (SH3DCPs) have a complex array of potential physiological partners due to their ability to recognize diverse structural scaffolds that are both dependent on, and independent of, the consensus proline-rich motif (PRM). This allows them to favor typical and atypical specific recognition sites. Biochemical and structural studies have been published on peptide libraries recognized by SH3 domains. These studies have been used to predict potential binding partners containing this sequence to gain a better understanding of SH3-mediated biological responses [10]. Human SH3DCPs represent a populous and well-characterized family with almost 300 domains embedded in 221 large multidomains and small monodomain proteins. A novel multidomain phylogenetic analysis of SH3DCPs shows their co-occurrence across a large set of protein domains, and it provides insight into their functional prerequisites in different signaling pathways.
2. Phylogenetic Classification of SH3DCPs
The next question researchers addressed concerned how to classify or categorize SH3DCPs, taking into account their heterogeneous domain composition. As the phylogenetic tree based on similarities of isolated SH3 domains was not of practical use, researchers have used an approach based on the similarities of domain compositions between SH3DCPs. For this purpose, primary sequences of an entire collection of 221 human SH3DCPs were first retrieved from the UniProt database, and they were analyzed for occurrences of protein domains. Next, mutual similarities in terms of domain composition between all protein pairs in the collection were evaluated. The resultant matrix was then subjected to phylogenetic analysis using MEGA software (version 7.0). The final phylogenic tree shed light on the evolutionary relationships between the human SH3DCP superfamily, and it allowed the superfamily to be classified into thirteen different SH3DCP families (Figure 2). An inspection of individual families, based on the respective domain organizations, revealed the following findings. (i) They differ in terms of the number of SH3DCPs per family, ranging from 2 (family 6) to 54 (family 3). (ii) The classification of SH3DCPs into individual families is often based on the combination of the SH3 domain with at least one or two similar domains, for example, SH2 and/or KinYST domains (Family 1); membrane-binding BAR and PH domains, RHOGAP, or RHOGEF domains (Family 2); single or several SH3 domains combined with other shared domains (Family 3); PDZ and/or the GuaKin domain (Family 4); FCH and/or RHOGAP domains (Family 5); UBA and HPhos domains (Family 6), S-rich and CAS-C domains (Family 7); Myosin and/or MyTH4 (Family 8); SAM* along with PTB and SLY in some SH3CPs (Family 9); spectrin domain and EF-hand (Family 11); DOCK and DHR domains (Family 12); and some were also classified with only a single SH3 domain (Family 13), except Family 10, which comprises diverse combinations of the SH3 domain. (iii) Exploiting combinations of SH3, with specific domains in each family of the SH3CPs’ domain–organization, indicates that the parallel domain–combination is evolving. This also explains the functional differentiation of the SH3 domain in different pathways. (iv) SH3 domains can function as adaptors, scaffolds, modulators, and regulatory domains.
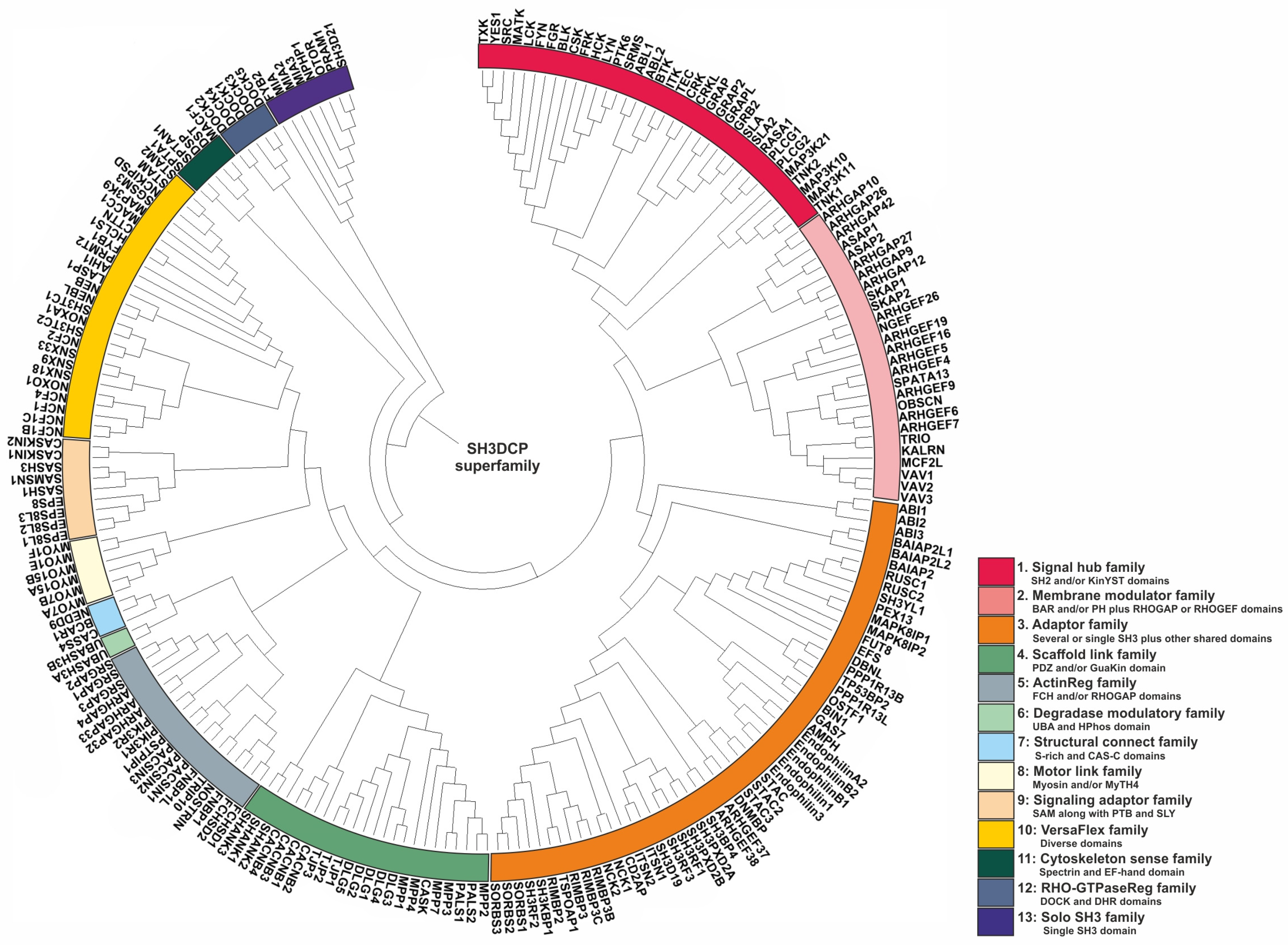
Figure 2. Phylogenetic tree of the SH3DCP superfamily. The tree was generated on the basis of the similarities between domain compositions. The SH3DCP superfamily can be divided into thirteen families, which are marked by different colors of classes at the outer ring.
2.1. Family 1
Proteins belonging to Family 1 share a mostly conserved domain called the tyrosine kinase domain, which is responsible for their catalytic activity and phosphorylation of target proteins. They can be classified into four groups of non-receptor Tyrosine Kinases (SRC, FYN, YES, HCK, LCK, BLK, FGR, FRK, SRMS, BTK, ITK, TEC, TXK, ABL1, ABL2, MATK, CSK, LYN, PTK6, TNK2, TNK1 [11][12]), adaptor Proteins (GRB2, GRAP, GRAP2, GRAPL, CRK, CRKL, SLA, SLA2 [13][14][15][16]), tyrosine Kinase-associated Signaling Proteins (RASA1, MAP3K21, MAP3K10, MAP3K11 [17][18], and Phospholipase C, including PLCG1 and PLCG2 [19][20]). The main feature of these proteins is that they are all involved in signal transduction pathways. More specifically, when transmitting signals from the cell surface to the cytoplasm and nucleus, they can affect gene expression and various cellular processes. The SH3 domain plays a crucial mediating interaction-based and regulatory role in this family. For example, SH3 domains of adaptor proteins, such as GRB2 and CRK, bind to proline-rich motifs in other signaling proteins, allowing them to link receptor tyrosine kinases to downstream signaling pathways [21][22]. In some cases, the SH3 domain affects the catalytic activity of the kinase domain. For instance, the SH3 domain of the non-receptor tyrosine kinase, SRC, can interact with its own SH2 domain and N-terminal fragment of the kinase domain, leading to the inactivation of its kinase activity [23].
2.2. Family 2
The proteins listed in Family 2 are primarily involved in the regulation of Rho family GTPases, and in some cases, those of the ARF family, which are critical for regulating the actin cytoskeleton and an array of essential cellular processes; these encompass cell migration, cell division, cell adhesion, and membrane trafficking. They can be classified further into two subcategories: GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs). GAPs are negative regulators of Rho or ARF family GTPases, and they stimulate the intrinsic GTPase activity of GTPases, which leads to their inactivation. The proteins of this family are GAPs, as follows: ARHGAP10, ARHGAP26, ARHGAP42, ARHGAP12, ARHGAP27, ARHGAP9 as RHOGAPs, and ASAP1, ASAP2 are ARF GAPs [24][25][26][27]. GEFs, on the other hand, activate GTPases by promoting the exchange of GDP for GTP. The proteins in the list are Rho-GEFs, as follows: SPATA13, ARHGEF4, ARHGEF26, NGEF, ARHGEF19, ARHGEF16, ARHGEF5, ARHGEF9, ARHGEF6, ARHGEF7 [25][28][29][30][31]. TRIO, KALRN, MCF2L, VAV1, VAV2, and VAV3 are multi-domain GEFs that regulate Rho family GTPases and other signaling pathways [25][32][33]. TRIO and KALRN activate RHO GTPases, RAC1 and RHOA, and they are involved in cell migration and differentiation [34]. VAV proteins and MCF2L activate RAC1, RHOA, and CDC42, and they are involved in cell growth, differentiation, and immune responses [32][35]. SKAP1 and SKAP2 do not have a canonical guanine nucleotide exchange factor (GEF) domain. Instead, they have been shown to act as RAP1 GTPase activators through a non-canonical mechanism that involves interactions with other proteins. More specifically, SKAP1 has been shown to bind to RIAM (RAP1-interacting adapter molecule), which, in turn, recruits activated GTP-bound RAP1 by promoting the membrane translocation of RAP1 for T-cell adhesion [36][37]. SKAP2 might also interact with RIAM, and it can similarly activate RAP1. Therefore, although SKAP1 and SKAP2 do not have a canonical GEF domain, they function as GEFs for RAP1 through protein–protein interactions with RIAM. There is a possibility that SH3 domains, similarly to other enzymes, control the activity of the GEF and GAP domains through inter/intra-molecular interactions. For example, unique characteristics were observed in this KALRN (kalirin) SH3 domain, including the presence of novel binding sites for the intramolecular PxxP ligand, as well as for binding to the adaptor protein, CRK, to inhibit the GEF activity of KALRN [38].
2.3. Family 3
The presence of multiple SH3 domains, in most members of Family 3, may confer several advantages, including increased specificity. First, having multiple SH3 domains with different binding specificities allows proteins to interact with a larger number of partner proteins and potentially simultaneously modulate multiple signaling pathways. Second, it might lead to cooperative binding, which means that the presence of multiple SH3 domains can allow a protein to bind to multiple sites on a single partner protein, which can enhance the affinity of the interaction and potentially stabilize protein complexes. A study conducted on a SH3RF3 protein from this family used a detailed functional scaffolding analysis that revealed that its fourth SH3 domain interacts with MKK7. Additionally, it was found that the first and second SH3 domains of SH3RF3 interact with JIP3 and JNK1. These findings suggest that SH3RF3 plays an important part in aiding the assembly of the MKK–JNK complex via JIP, which leads to the activation of JNK-JUN [39]. Thirdly, the regulation of protein–protein interactions occurs when the SH3 domains in a protein can interact with each other, or with other domains within the same protein, to regulate protein–protein interactions. For example, autoinhibitory interactions of SH3 domains can block binding sites and prevent interactions until a regulatory signal is received. For example, ITSN1-L, which is a RHO-GEF, plays a crucial role in regulating both endocytosis and actin cytoskeletal rearrangements, and its SH3 domains are important for controlling its exchange activity. The SH3 domains block the binding of CDC42 to the RHO-GEF domain (or DH domain) via inter-domain interactions, which inhibits exchange activity [40]. Lastly, localization concerns the presence of multiple SH3 domains with different binding specificities, which can also allow proteins to target different subcellular compartments and interact with different sets of proteins in those locations. Interestingly, the specific order and arrangement of the SH3 domains were found to be important for maintaining the integrity of protein–protein networks in SH3CPs with multiple SH3 domains [6].
Members of this family can also function as adaptor proteins that typically contain multiple domains, and they can couple together different signaling molecules or components of cellular pathways. Many proteins of Family 3 (SH3CPs) fall into this category. For example, CD2AP (CD2-associated protein) is an adaptor protein that interacts with CD2, a transmembrane receptor protein on T cells [41], and other cytosolic proteins such as nephrin, a protein important for maintaining the integrity of the glomerular filtration barrier in the kidney [42]. The SH3 domains in CD2AP are thought to mediate protein–protein interactions with other signaling molecules and cytoskeletal components [43]. NCK1 and NCK2 (non-catalytic region of tyrosine kinase) proteins are adaptor proteins that link signaling molecules with downstream effector proteins involved in cytoskeletal regulation, membrane trafficking, and gene expression [43][44]. They contain several protein-binding domains, including SH3 domains, that enable them to simultaneously interact with multiple partners. In addition, all RIMBP proteins (RIMBP2, RIMBP3, RIMBP3B, RIMBP3C) are part of the synaptic vesicle release machinery and are involved in regulating neurotransmitter release [45]. They contain several domains that allow them to interact with other proteins involved in the synaptic vesicle cycle. Another category that some members of this family fall into is signaling proteins that act as intermediates or effectors in various signaling pathways. Some examples of MAPK8IP1 and MAPK8IP2 are as follows. MAPK8IP proteins (mitogen-activated protein kinase 8 interacting protein) are involved in the regulation of the JNK (c-Jun N-terminal kinase) signaling pathway, which is important for stress responses and apoptosis [46]. The SH3 domain in these proteins mediates protein–protein interactions with upstream and downstream components of the pathway. STAC, STAC2, and STAC3 are types of STAC protein that are involved in the regulation of calcium channels, and they play a role in skeletal muscle function. They contain several domains, including the SH3 domain, that interact with different components of the calcium channel complex [47]. OSTF1 (Osteoclast-stimulating factor 1) is another example of the protein involved in the regulation of bone resorption by osteoclasts [48]. The SH3 domain in OSTF1 is thought to mediate interactions using signaling molecules involved in the regulation of osteoclast activity. SH3 domain-containing cytoskeletal proteins, categorized as cytoskeletal proteins, such as Endophilins (Endophilin A2, Endophilin B1, Endophilin B2, Endophilin 1, Endophilin 3), are involved with controlling the organization and dynamics of the cell cytoskeleton. They are involved in the formation and recycling of clathrin-coated vesicles and the regulation of the actin cytoskeleton. Endophilins contain, among others, a BAR domain which further contributes to their membrane curvature recognition [49]. They also interact with proteins such as dynamin and synaptojanin via the SH3 domain which regulates the formation of clathrin-coated vesicles during endocytosis [50][51]. DNMBP or TUBA (Dynamin-binding protein) is also involved in actin cytoskeleton organization, and it is thought to play a role in endocytosis. The SH3 domains in DNMBP are involved in protein–protein interactions with other cytoskeletal and signaling proteins [52].
2.4. Family 4
Proteins listed in this family share SH3 domains and/or PDZ and/or Guanylate Kinase (GuaKin/GK) domains, and they often have similar functions associated with the regulation of protein complexes and the structure and function of the synapse, a junction between two neurons that allows for the transmission of information. SHANK1, SHANK2, and SHANK3 are scaffolding proteins that play a crucial role in the organization and function of the postsynaptic density (PSD), a protein-rich area of the synapse [53]. SHANK proteins interact with other proteins to anchor neurotransmitter receptors and signaling molecules in the PSD, thereby regulating the strength of synaptic transmission [54]. MPP1, MPP2, MPP3, MPP4, MPP7, PALS1, and PALS2 are members of the membrane-associated guanylate kinase (MAGUK) family in synapse organization and function. MAGUK proteins interact with other proteins to form a complex network of signaling molecules at the synapse, thereby regulating synaptic transmission and plasticity [55][56]. CASK is a protein in the same subfamily that interacts with other synaptic proteins, including β-neurexins, and Rabphilin3a via the PDZ domain; it plays a role in the regulation of neurotransmitter release [55]. DLG1, DLG2, DLG3, DLG4, and DLG5 are members of the Discs Large (DLG) subfamily of proteins belonging to the MAGUK family; they are involved in the formation and control of neurotransmitter release [57][58]. CACNB1, CACNB2, CACNB3, and CACNB4 are subunits of voltage-gated calcium channels (VGCCs) belonging to the MAGUK family; they regulate the entry of calcium ions into neurons. Calcium influx through VGCCs is important for synaptic plasticity and neurotransmitter release [59]. In contrast, TJP1, TJP2, and TJP3, which belong to the ZO subfamily, are also members of the MAGUK family. They are not expressed in neurons, but in the brain, and they play a crucial role in maintaining the blood–brain barrier [60].
Overall, although the SH3 domain’s interaction with its targets is less understood compared with PDZ domains, studies on MAGUK proteins, such as DLG, provide insights into the complex regulation of SH3 domain interactions and their potential roles in cellular processes. The N-terminal region of the human DLG undergoes alternative proline-rich region insertion splicing that can bind in vitro to multiple SH3 domains and control the formation of protein clusters [61]. For example, the N-terminal portion of DLG1 (SAP-97) can bind to the SH3 segment of DLG4 (PSD-95), indicating a potential heteromeric interaction between these two proteins. This interaction may play a role in dendritic clustering and the trafficking of GluR-A AMPA receptors [62]. Other studies suggest that the SH3 domain of DLG1 (SAP-97) and DLG4 (PSD-95) forms a specific interaction with its GK domain, and this intramolecular interaction prevents intermolecular associations; this sheds light on the role of the SH3 domain with regard to MAGUK function and oligomerization [63][64][65]. Recent findings suggest that the SH3 domain modulates the GK domain through an allosteric mechanism rather than by blocking the GK binding surface [66]. The SH3-HOOK-GK domain configuration is present in most MAGUK proteins, suggesting that this interaction is a shared characteristic among MAGUK proteins [64][67]. Overall, these proteins play important roles in regulating the organization and function of the synapse, and the dysregulation of their activity has been linked to various neurological and psychiatric disorders.
2.5. Family 5
Proteins classified into this family share a similar domain architecture. They all contain at least one SH3 domain, either a FCH or RHOGAP domain, or both. This combination of domains is unique to this protein family and sets them apart from other proteins. The combination of the SH3 domain with the FCH and/or RHOGAP domains in this group of proteins suggests that they may play a role in regulating actin cytoskeleton dynamics and membrane trafficking. Proteins containing the FCH domain may participate in protein–protein interactions, and they may potentially contribute to the organization of RHO proteins and the actin cytoskeleton [68]. Conversely, the RHOGAP domain regulates RHO family GTPases, which are important regulators of actin cytoskeleton dynamics.
2.6. Family 6
UBASH3A and UBASH3B are two proteins that belong to the same protein family, called the Ubiquitin-associated and SH3 domain-containing protein (UBASH3) family. These proteins are involved in the regulation of signal transduction pathways, including T-cell receptor signaling and cytokine production [69][70][71]. Functionally, both UBASH3A and UBASH3B contain an Ubiquitin-associated (UBA) domain and a SRC homology 3 (SH3) domain. The UBA domain enables the interaction between these proteins and ubiquitin, a protein that plays a critical role in the regulation of protein degradation, DNA repair, and immune response [72]. The SH3 domain allows UBASH3A and UBASH3B to bind to proline-rich motifs in other proteins, including signaling proteins, receptors, and enzymes, thereby regulating their activity [70][73][74]. Structurally, UBASH3A and UBASH3B are similar in size, each consisting of 504 amino acid residues. Both proteins share a high degree of sequence identity, with 80% sequence similarity. The overall structure of these proteins is similar, with an N-terminal UBA domain followed by a central SH3 domain and a C-terminal HPhos region. However, there are some differences in the sequence and structure of the UBA and SH3 domains between UBASH3A and UBASH3B, which may contribute to their distinct functions.
2.7. Family 7
BCAR1, NEDD9, and CASS4 also share other domains in addition to the SH3 domain, namely, the S-RICH and CAS-C domains. The S-RICH domain, which is a stretch of amino acids enriched with serine residues, is located in the N-terminal region of all three proteins. It has been shown to be crucial for the localization and activity of these proteins at focal adhesions, which are sites of cell adhesion and signaling, by binding to 14-3-3 proteins [75]. The CAS-C domain is a domain that is found in the C-terminal region of all three proteins, and it assists with binding to other signaling molecules, such as the adapter protein, SRC, which mediates downstream signaling events [76][77]. Therefore, the common structural feature of the SH3 domain, coupled with the S-RICH and CAS-C domains, contributes to the functional similarities between BCAR1, NEDD9, and CASS4; this is because it allows them to interact with the other proteins involved in cell adhesion and signaling pathways, leading to similar functional roles in terms of regulating cell adhesion, migration, and proliferation [76].
2.8. Family 8
These proteins share common structural features in that they all contain SH3 with myosin domains belonging to the myosin superfamily. Myosins are a family of motor proteins that use the energy from ATP hydrolysis to generate force and move along actin filaments, resulting in the generation of force and motion [78]. Functionally, myosins are involved in a wide range of cellular processes, including muscle contraction, cell migration, membrane trafficking, and organelle transport [79]. The specific functions of listed myosins may vary depending on their expression patterns, subcellular localization, and interactions with other proteins. For example, MYO7A is involved in hearing and balance [80], whereas MYO5A is involved in melanosome transport and pigmentation [81]. Other myosins, such as MYO1E, are involved in cell migration and the regulation of the actin cytoskeleton [82]. Myosins can be divided into two broad categories, as follows: conventional and unconventional myosins. Conventional myosins are typically found in muscle tissue and are responsible for generating the force and movement required for muscle contraction [83]. Unconventional myosins, on the other hand, have a more diverse range of functions, and they are found in a variety of cell types and tissues throughout the body [84]. Some examples of these unconventional roles include acting as tension sensors and dynamic tethers, organizing F-actin during endo- and exocytosis, and maintaining the mitotic spindle structure [85]. Unconventional myosins often have a more complex domain structure than conventional myosins, and the SH3 domain is one of the additional domains that is commonly found in these proteins [84]. The SH3 domain in some myosins, given their interaction with other proteins, may carry out these functions [86]. All listed myosins contain a single SH3 domain, which is involved in mediating protein–protein interactions, and this is consistent with the idea that these myosins play roles in diverse unconventional processes. For example, MYO1E, which is an unconventional myosin involved in the cell migration and regulation of the actin cytoskeleton, contains a SH3 domain that has been shown to interact with a protein called ZO-1 [82]. This interaction is thought to play a role in regulating junctional integrity in kidney podocytes by contributing to the slit diaphragm complex [87]. Similarly, MYO7A, which is involved in hearing and balance, contains an SH3 domain that contributes to the interaction with the protein harmonin. This interaction is important for the localization of MYO7A to the stereocilia in the inner ear, where it is involved in generating mechanical force and movement [88].
2.9. Family 9
The concurrent presence of SH3 domains and SAM*, PTB, and SLY domains in some of the proteins listed in Family 9 suggests that they play roles in various aspects of signal transduction and protein–protein interactions. The SAM* (sterile alpha motif) domain is a conserved protein domain of around 70 amino acids that is present in many proteins involved in signal transduction and transcriptional regulation [89]. SAM* domains are known to mediate protein–protein interactions and are believed to function as regulatory domains that can influence the activity or localization of their associated proteins [90]. The PTB (phosphotyrosine binding) domain is another protein domain that is commonly found in signaling proteins. PTB domains bind to specific phosphorylated tyrosine residues in other proteins, and they are involved in mediating protein–protein interactions that are essential for the proper functioning of signaling pathways [91]. The SLY domain, a conserved family of lymphocyte signaling adapter proteins domain, is present in eukaryotes and is associated with SH3 and SAM domains. It is identified in various proteins, including SLY1/SASH1, SASH3, and SAMSN1 [92]. The combined presence of these domains in listed proteins suggests that they likely function as adaptors or scaffold proteins that help to assemble and organize signaling complexes, and that they mediate the protein–protein interactions that are critical for signaling and regulation. Adaptor proteins contain protein–protein interaction domains that link receptors to downstream signaling components, whereas scaffold proteins provide a physical platform for multiple signaling components to interact with and regulate each other’s activity. Based on their known functions and structural features, EPS8, EPS8L1-3 [93], SASH1 [94], SASH3, and SAMSN1 [95] are believed to function as adaptor proteins, whereas CASKIN1 and CASKIN2 are scaffold proteins. CASKIN1 and CASKIN2 contain multiple domains which enable them to function as scaffold proteins that can organize multi-protein complexes [96]. The structural investigation of CASKIN2′s SH3 domain using NMR revealed that its peptide-binding cleft differed from the typical binding sites for polyproline ligands due to the presence of non-canonical basic amino acids. Mutations in the cleft suggested that the SH3 domain in CASKIN2 may have lost its functional ability to promote protein–protein interactions beyond the conventional roles typically associated with SH3 domains [97].
2.10. Family 10
Although SH3 domains may be a shared feature among these proteins, their overall domain architectures and functions are diverse. Therefore, it is important to note that some of these proteins may have multiple functions, or they may interact with multiple signaling pathways; their precise classification can depend on context and experimental findings. However, they can be primarily classified into the following functional categories: signal transduction (STAM, STAM2 [98], NCKIPSD [99], MAP3K9 [100], MACC1 [101], PRMT2 [102], AHI1 [103], LASP1 [104], SGSM3 [105]), cytoskeletal remodeling (HCLS1 [106], CTTN [107], NEBL, NEB [108], LASP1 [104], FYB [109]), endocytosis (SH3TC1, SH3TC2 [110], SNX9, SNX33, SNX18 [111]), and immune system function (NCF1, NCF1B, NCF1C, NCF2, NCF4 [112], NOXO1, NOXA1 [113]). Furthermore, many of these proteins have multiple SH3 domains, and some may have other protein–protein interaction domains or motifs that contribute to their functions.
2.11. Family 11
The shared structural and functional features of these proteins are primarily related to their roles in cytoskeletal organization and cell adhesion. The spectrin domain is a key structural component that provides mechanical stability to the cytoskeleton. It forms a long, flexible rod-like structure that can interact with other proteins, cytoskeletal elements, and lipids to provide support and resistance against deformation [114][115][116]. The SH3 domain, on the other hand, plays a key role in cytoskeletal organization and cell adhesion by regulating protein–protein interactions and localization. The EF-hands have a high affinity for Ca2+, they undergo a conformational change when bound to it, and they are essential for maintaining the structural integrity of the skeleton [115]. Together, the SH3, spectrin, and EF-hand domains found in these proteins can work together to regulate critical protein–protein interactions that maintain the structural integrity of the cytoskeleton and regulate cellular adhesion and signaling. Although each of these proteins have unique features and functions, they all share common structural and functional elements that reflect their common ancestry and evolutionary history.
2.12. Family 12
These proteins share both structural and functional similarities as they all belong to the same family of guanine nucleotide exchange factors (GEFs), known as the DOCK family. Structurally, they all contain a conserved DHR-2-C (DOCK homology region 2) domain which is responsible for the GEF activity of these proteins, as well as other domains such as DHR-2-A (lipid-binding DOCK homology region) and the SH3 domain. Functionally, they play important roles in the regulation of cytoskeletal dynamics, cell migration, and immune and neural cell function [117][118]. In the DOCK family, the SH3 domain plays a regulatory role by mediating interactions with proline-rich motifs in other proteins, allowing DOCK proteins to bind to, and regulate the activity of, a variety of cytoskeletal and signaling proteins. There are some examples of how the SH3 domain in DOCK proteins can play a role in regulating protein–protein interactions. The SH3 domain of DOCK2 interacts with the PRM of ELMO1, which may relieve their autoinhibition to promote the activation of RAC in lymphocyte chemotaxis [119][120]. Moreover, the DOCK1–ELMO1 interaction was identified for the localization and regulation of RAC1 in cytoskeletal organization and cell migration [120][121]. The C-terminal PRM region of DOCK1 can also interact with the SH3 domain of several proteins, including the adaptor protein, NCKβ, and CRK, which helps to control cell migration [122][123][124]. Thus, the SH3 domain is an important structural component that facilitates these interactions to influence the subcellular activity of DOCK proteins, as well as their ability to activate downstream signaling pathways.
2.13. Family 13
All of these proteins contain only one SH3 domain. The specific function of each protein may be different, but they all share the ability to interact with other proteins via their SH3 domain. For example, FYB2 (FYN binding protein 2) regulates T-cell receptor signaling and is involved in the formation of the immunological synapse [125]. Another study found that MIA, a protein secreted from malignant melanoma cells, enhances melanoma cell migration and invasion by interacting with extracellular matrix proteins and integrin [126][127]. In addition, cadherin-7 was identified as a new MIA-binding protein that negatively regulates the expression and activity of MIA, and it plays a role in the migration of melanoma cells during tumor development [128]. Another review integrates research on Drosophila Tango1 and human MIA/cTAGE proteins to provide an evolutionary perspective on ER-Golgi transport, which highlights the role of the MIA protein involved in the regulation of the ER-Golgi transport of proteins [129]. OTOR (melanoma inhibitory activity-like (alias MIAL)) may play a role in the development and maintenance of the inner ear [130]. NPHP1 (Nephrocystin-1) plays a role in the macromolecular complex formation and function of cilia, and disruptions to these complexes can cause renal cystogenesis [131]. PRAM (PML-RAR alpha-regulated adapter molecule) is involved in the regulation of the differentiation of hematopoietic cells [132]. SH3D21′s (SH3 domain-containing protein 21) function is currently unknown, and further research is needed to fully understand the specific role and mechanisms of SH3D21 with regard to signaling processes. The cellular localization of these proteins may vary depending on their specific function and the cell type in which they are expressed. Although some proteins may have a predominant localization to a particular subcellular compartment, others may be distributed more broadly throughout the cell.
This entry is adapted from the peer-reviewed paper 10.3390/cells12162054
References
- Mayer, B.J.; Hamaguchi, M.; Hanafusa, H. A novel viral oncogene with structural similarity to phospholipase C. Nature 1988, 332, 272.
- Stahl, M.L.; Ferenz, C.R.; Kelleher, K.L.; Kriz, R.W.; Knopf, J.L. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature 1988, 332, 269–272.
- Musacchio, A.; Noble, M.; Pauptit, R.; Wierenga, R.; Saraste, M. Crystal structure of a Src-homology 3 (SH3) domain. Nature 1992, 359, 851–855.
- Gmeiner, W.H.; Horita, D.A. Implications of SH3 domain structure and dynamics for protein regulation and drug design. Cell Biochem. Biophys. 2001, 35, 127–140.
- Whisstock, J.C.; Lesk, A.M. SH3 domains in prokaryotes. Trends Biochem. Sci. 1999, 24, 132–133.
- Dionne, U.; Bourgault, É.; Dubé, A.K.; Bradley, D.; Chartier, F.J.M.; Dandage, R.; Dibyachintan, S.; Després, P.C.; Gish, G.D.; Pham, N.T.H.; et al. Protein context shapes the specificity of SH3 domain-mediated interactions in vivo. Nat. Commun. 2021, 12, 1597.
- Morel, B.; Varela, L.; Azuaga, A.I.; Conejero-Lara, F. Environmental conditions affect the kinetics of nucleation of amyloid fibrils and determine their morphology. Biophys. J. 2010, 99, 3801–3810.
- Smithgall, T.E. SH2 and SH3 domains: Potential targets for anti-cancer drug design. J. Pharmacol. Toxicol. Methods 1995, 34, 125–132.
- Kadaveru, K.; Vyas, J.; Schiller, M.R. Viral infection and human disease--insights from minimotifs. Front. Biosci. 2008, 13, 6455–6471.
- Teyra, J.; Huang, H.; Jain, S.; Guan, X.; Dong, A.; Liu, Y.; Tempel, W.; Min, J.; Tong, Y.; Kim, P.M.; et al. Comprehensive Analysis of the Human SH3 Domain Family Reveals a Wide Variety of Non-canonical Specificities. Structure 2017, 25, 1598–1610.e1593.
- Siveen, K.S.; Prabhu, K.S.; Achkar, I.W.; Kuttikrishnan, S.; Shyam, S.; Khan, A.Q.; Merhi, M.; Dermime, S.; Uddin, S. Role of Non Receptor Tyrosine Kinases in Hematological Malignances and its Targeting by Natural Products. Mol. Cancer 2018, 17, 31.
- Mahajan, K.; Mahajan, N.P. PI3K-independent AKT activation in cancers: A treasure trove for novel therapeutics. J. Cell. Physiol. 2012, 227, 3178–3184.
- Pawson, T. Specificity in Signal Transduction: From Phosphotyrosine-SH2 Domain Interactions to Complex Cellular Systems. Cell 2004, 116, 191–203.
- Lundby, A.; Franciosa, G.; Emdal, K.B.; Refsgaard, J.C.; Gnosa, S.P.; Bekker-Jensen, D.B.; Secher, A.; Maurya, S.R.; Paul, I.; Mendez, B.L.; et al. Oncogenic Mutations Rewire Signaling Pathways by Switching Protein Recruitment to Phosphotyrosine Sites. Cell 2019, 179, 543–560.e526.
- Birge, R.B.; Kalodimos, C.; Inagaki, F.; Tanaka, S. Crk and CrkL adaptor proteins: Networks for physiological and pathological signaling. Cell Commun. Signal. 2009, 7, 13.
- Moharram, S.A.; Rönnstrand, L.; Kazi, J.U. Src-Like Adapter Protein (SLAP). In Encyclopedia of Signaling Molecules; Choi, S., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 5145–5149.
- Pamonsinlapatham, P.; Hadj-Slimane, R.; Lepelletier, Y.; Allain, B.; Toccafondi, M.; Garbay, C.; Raynaud, F. P120-Ras GTPase activating protein (RasGAP): A multi-interacting protein in downstream signaling. Biochimie 2009, 91, 320–328.
- Chadee, D.N. Involvement of mixed lineage kinase 3 in cancer. Can. J. Physiol. Pharmacol. 2013, 91, 268–274.
- Kim, H.Y.; Suh, P.-G.; Kim, J.-I. The Role of Phospholipase C in GABAergic Inhibition and Its Relevance to Epilepsy. Int. J. Mol. Sci. 2021, 22, 3149.
- Emmanouilidi, A.; Lattanzio, R.; Sala, G.; Piantelli, M.; Falasca, M. The role of phospholipase Cγ1 in breast cancer and its clinical significance. Future Oncol. 2017, 13, 1991–1997.
- Kazemein Jasemi, N.S.; Herrmann, C.; Magdalena Estirado, E.; Gremer, L.; Willbold, D.; Brunsveld, L.; Dvorsky, R.; Ahmadian, M.R. The intramolecular allostery of GRB2 governing its interaction with SOS1 is modulated by phosphotyrosine ligands. Biochem. J. 2021, 478, 2793–2809.
- Schumacher, C.; Knudsen, B.S.; Ohuchi, T.; Fiore, P.P.D.; Glassman, R.H.; Hanafusa, H. The SH3 Domain of Crk Binds Specifically to a Conserved Proline-rich Motif in Eps15 and Eps15R (∗). J. Biol. Chem. 1995, 270, 15341–15347.
- Tatosyan, A.; Mizenina, O. Kinases of the Src family: Structure and functions. BIOCHEMISTRY C/C OF BIOKHIMIIA 2000, 65, 49–58.
- Santy, L.C.; Casanova, J.E. GTPase Signaling: Bridging the GAP between ARF and Rho. Curr. Biol. 2002, 12, R360–R362.
- Vigil, D.; Cherfils, J.; Rossman, K.L.; Der, C.J. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 2010, 10, 842–857.
- Furukawa, Y.; Kawasoe, T.; Daigo, Y.; Nishiwaki, T.; Ishiguro, H.; Takahashi, M.; Kitayama, J.; Nakamura, Y. Isolation of a novel human gene, ARHGAP9, encoding a rho-GTPase activating protein. Biochem. Biophys. Res. Commun. 2001, 284, 643–649.
- Sakakibara, T.; Nemoto, Y.; Nukiwa, T.; Takeshima, H. Identification and characterization of a novel Rho GTPase activating protein implicated in receptor-mediated endocytosis. FEBS Lett. 2004, 566, 294–300.
- Bourgeois, J.S.; Wang, L.; Rabino, A.F.; Everitt, J.; Alvarez, M.I.; Awadia, S.; Wittchen, E.S.; Garcia-Mata, R.; Ko, D.C. ARHGEF26 enhances Salmonella invasion and inflammation in cells and mice. PLoS Pathog. 2021, 17, e1009713.
- Zhang, M.; Lin, L.; Wang, C.; Zhu, J. Double inhibition and activation mechanisms of Ephexin family RhoGEFs. Proc. Natl. Acad. Sci. 2021, 118, e2024465118.
- Zamboni, V.; Jones, R.; Umbach, A.; Ammoni, A.; Passafaro, M.; Hirsch, E.; Merlo, G.R. Rho GTPases in intellectual disability: From genetics to therapeutic opportunities. Int. J. Mol. Sci. 2018, 19, 1821.
- López Tobón, A.; Suresh, M.; Jin, J.; Vitriolo, A.; Pietralla, T.; Tedford, K.; Bossenz, M.; Mahnken, K.; Kiefer, F.; Testa, G.; et al. The guanine nucleotide exchange factor Arhgef7/βPix promotes axon formation upstream of TC10. Sci. Rep. 2018, 8, 8811.
- Maiwald, S.; Motazacker, M.M.; van Capelleveen, J.C.; Sivapalaratnam, S.; van der Wal, A.C.; van der Loos, C.; Kastelein, J.J.; Ouwehand, W.H.; Hovingh, G.K.; Trip, M.D.; et al. A rare variant in MCF2L identified using exclusion linkage in a pedigree with premature atherosclerosis. Eur. J. Hum. Genet. 2016, 24, 86–91.
- Paskus, J.D.; Herring, B.E.; Roche, K.W. Kalirin and Trio: RhoGEFs in synaptic transmission, plasticity, and complex brain disorders. Trends Neurosci. 2020, 43, 505–518.
- Grubisha, M.J.; DeGiosio, R.A.; Wills, Z.P.; Sweet, R.A. Trio and Kalirin as unique enactors of Rho/Rac spatiotemporal precision. Cell Signal 2022, 98, 110416.
- Abe, K.; Rossman, K.L.; Liu, B.; Ritola, K.D.; Chiang, D.; Campbell, S.L.; Burridge, K.; Der, C.J. Vav2 is an activator of Cdc42, Rac1, and RhoA. J. Biol. Chem. 2000, 275, 10141–10149.
- Kliche, S.; Breitling, D.; Togni, M.; Pusch, R.; Heuer, K.; Wang, X.; Freund, C.; Kasirer-Friede, A.; Menasche, G.; Koretzky, G.A.; et al. The ADAP/SKAP55 signaling module regulates T-cell receptor-mediated integrin activation through plasma membrane targeting of Rap1. Mol. Cell Biol. 2006, 26, 7130–7144.
- Ménasché, G.; Kliche, S.; Chen, E.J.; Stradal, T.E.; Schraven, B.; Koretzky, G. RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation. Mol. Cell Biol. 2007, 27, 4070–4081.
- Schiller, M.R.; Chakrabarti, K.; King, G.F.; Schiller, N.I.; Eipper, B.A.; Maciejewski, M.W. Regulation of RhoGEF activity by intramolecular and intermolecular SH3 domain interactions. J. Biol. Chem. 2006, 281, 18774–18786.
- Zhang, P.; Liu, Y.; Lian, C.; Cao, X.; Wang, Y.; Li, X.; Cong, M.; Tian, P.; Zhang, X.; Wei, G. SH3RF3 promotes breast cancer stem-like properties via JNK activation and PTX3 upregulation. Nat. Commun. 2020, 11, 1–13.
- Zamanian, J.L.; Kelly, R.B. Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent src homology 3 domains that are also involved in endocytosis. Mol. Biol. Cell 2003, 14, 1624–1637.
- Binder, C.; Cvetkovski, F.; Sellberg, F.; Berg, S.; Paternina Visbal, H.; Sachs, D.H.; Berglund, E.; Berglund, D. CD2 immunobiology. Front. Immunol. 2020, 11, 1090.
- Martin, C.E.; Jones, N. Nephrin signaling in the podocyte: An updated view of signal regulation at the slit diaphragm and beyond. Front. Endocrinol. 2018, 9, 302.
- Machuca, E.; Benoit, G.; Antignac, C. Genetics of nephrotic syndrome: Connecting molecular genetics to podocyte physiology. Hum. Mol. Genet. 2009, 18, R185–R194.
- Li, W.; Fan, J.; Woodley, D.T. Nck/Dock: An adapter between cell surface receptors and the actin cytoskeleton. Oncogene 2001, 20, 6403–6417.
- Brockmann, M.M.; Zarebidaki, F.; Camacho, M.; Grauel, M.K.; Trimbuch, T.; Südhof, T.C.; Rosenmund, C. A Trio of Active Zone Proteins Comprised of RIM-BPs, RIMs, and Munc13s Governs Neurotransmitter Release. Cell Rep. 2020, 32, 107960.
- Hammouda, M.B.; Ford, A.E.; Liu, Y.; Zhang, J.Y. The JNK signaling pathway in inflammatory skin disorders and cancer. Cells 2020, 9, 857.
- Flucher, B.E.; Campiglio, M. STAC proteins: The missing link in skeletal muscle EC coupling and new regulators of calcium channel function. Biochim. Et Biophys. Acta (BBA)—Mol. Cell Res. 2019, 1866, 1101–1110.
- Vermeren, M.; Lyraki, R.; Wani, S.; Airik, R.; Albagha, O.; Mort, R.; Hildebrandt, F.; Hurd, T. Osteoclast stimulation factor 1 (Ostf1) KNOCKOUT increases trabecular bone mass in mice. Mamm. Genome 2017, 28, 498–514.
- Milosevic, I.; Giovedi, S.; Lou, X.; Raimondi, A.; Collesi, C.; Shen, H.; Paradise, S.; O’Toole, E.; Ferguson, S.; Cremona, O.; et al. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron 2011, 72, 587–601.
- Ringstad, N.; Nemoto, Y.; De Camilli, P. Differential Expression of Endophilin 1 and 2 Dimers at Central Nervous System Synapses*. J. Biol. Chem. 2001, 276, 40424–40430.
- Cestra, G.; Castagnoli, L.; Dente, L.; Minenkova, O.; Petrelli, A.; Migone, N.; Hoffmüller, U.; Schneider-Mergener, J.; Cesareni, G. The SH3 Domains of Endophilin and Amphiphysin Bind to the Proline-rich Region of Synaptojanin 1 at Distinct Sites That Display an Unconventional Binding Specificity*. J. Biol. Chem. 1999, 274, 32001–32007.
- Salazar, M.A.; Kwiatkowski, A.V.; Pellegrini, L.; Cestra, G.; Butler, M.H.; Rossman, K.L.; Serna, D.M.; Sondek, J.; Gertler, F.B.; De Camilli, P. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem. 2003, 278, 49031–49043.
- Kreienkamp, H.-J. Scaffolding proteins at the postsynaptic density: Shank as the architectural framework. In Protein-Protein Interactions as New Drug Targets; Springer: Berlin/Heidelberg, Germany, 2008; pp. 365–380.
- Shi, R.; Redman, P.; Ghose, D.; Hwang, H.; Liu, Y.; Ren, X.; Ding, L.J.; Liu, M.; Jones, K.J.; Xu, W. Shank Proteins Differentially Regulate Synaptic Transmission. Eneuro 2017, 4.
- Oliva, C.; Escobedo, P.; Astorga, C.; Molina, C.; Sierralta, J. Role of the MAGUK protein family in synapse formation and function. Dev. Neurobiol. 2012, 72, 57–72.
- De Mendoza, A.; Suga, H.; Ruiz-Trillo, I. Evolution of the MAGUK protein gene family in premetazoan lineages. BMC Evol. Biol. 2010, 10, 93.
- Budnik, V.; Koh, Y.-H.; Guan, B.; Hartmann, B.; Hough, C.; Woods, D.; Gorczyca, M. Regulation of Synapse Structure and Function by the Drosophila Tumor Suppressor Gene dlg. Neuron 1996, 17, 627–640.
- Wilkinson, B.; Coba, M.P. Molecular architecture of postsynaptic Interactomes. Cell Signal 2020, 76, 109782.
- Rima, M.; Daghsni, M.; Fajloun, Z.; M’rad, R.; Brusés, J.L.; Ronjat, M.; De Waard, M. Protein partners of the calcium channel β subunit highlight new cellular functions. Biochem. J. 2016, 473, 1831–1844.
- Wolburg, H.; Lippoldt, A. Tight junctions of the blood–brain barrier: Development, composition and regulation. Vasc. Pharmacol. 2002, 38, 323–337.
- McLaughlin, M.; Hale, R.; Ellston, D.; Gaudet, S.; Lue, R.A.; Viel, A. The Distribution and Function of Alternatively Spliced Insertions in hDlg*. J. Biol. Chem. 2002, 277, 6406–6412.
- Cai, C.; Li, H.; Rivera, C.; Keinänen, K. Interaction between SAP97 and PSD-95, Two Maguk Proteins Involved in Synaptic Trafficking of AMPA Receptors*. J. Biol. Chem. 2006, 281, 4267–4273.
- McGee, A.W.; Bredt, D.S. Identification of an Intramolecular Interaction between the SH3 and Guanylate Kinase Domains of PSD-95*. J. Biol. Chem. 1999, 274, 17431–17436.
- McGee, A.W.; Dakoji, S.R.; Olsen, O.; Bredt, D.S.; Lim, W.A.; Prehoda, K.E. Structure of the SH3-Guanylate Kinase Module from PSD-95 Suggests a Mechanism for Regulated Assembly of MAGUK Scaffolding Proteins. Mol. Cell 2001, 8, 1291–1301.
- Wu, H.; Reissner, C.; Kuhlendahl, S.; Coblentz, B.; Reuver, S.; Kindler, S.; Gundelfinger, E.D.; Garner, C.C. Intramolecular interactions regulate SAP97 binding to GKAP. Embo J. 2000, 19, 5740–5751.
- Marcette, J.; Hood, I.V.; Johnston, C.A.; Doe, C.Q.; Prehoda, K.E. Allosteric control of regulated scaffolding in membrane-associated guanylate kinases. Biochemistry 2009, 48, 10014–10019.
- Tavares, G.A.; Panepucci, E.H.; Brunger, A.T. Structural Characterization of the Intramolecular Interaction between the SH3 and Guanylate Kinase Domains of PSD-95. Mol. Cell 2001, 8, 1313–1325.
- Aspenström, P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 1997, 7, 479–487.
- Wang, Z.; Wang, Y.; Peng, M.; Yi, L. UBASH3B is a novel prognostic biomarker and correlated with immune infiltrates in prostate cancer. Front. Oncol. 2020, 9, 1517.
- Ge, Y.; Paisie, T.; Chen, S.; Concannon, P. UBASH3A regulates the synthesis and dynamics of T-cell receptor-CD3 complexes. J. Immunol. 2019, 203, 2827–2836.
- Yamagata, K.; Nakayamada, S.; Zhang, T.; Nguyen, A.P.; Ohkubo, N.; Iwata, S.; Kato, S.; Tanaka, Y. IL-6 production through repression of UBASH3A gene via epigenetic dysregulation of super-enhancer in CD4+ T cells in rheumatoid arthritis. Inflamm. Regen. 2022, 42, 46.
- Hoeller, D.; Crosetto, N.; Blagoev, B.; Raiborg, C.; Tikkanen, R.; Wagner, S.; Kowanetz, K.; Breitling, R.; Mann, M.; Stenmark, H.; et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 2006, 8, 163–169.
- Feshchenko, E.A.; Smirnova, E.V.; Swaminathan, G.; Teckchandani, A.M.; Agrawal, R.; Band, H.; Zhang, X.; Annan, R.S.; Carr, S.A.; Tsygankov, A.Y. TULA: An SH3- and UBA-containing protein that binds to c-Cbl and ubiquitin. Oncogene 2004, 23, 4690–4706.
- Ge, Y.; Paisie, T.K.; Newman, J.R.B.; McIntyre, L.M.; Concannon, P. UBASH3A Mediates Risk for Type 1 Diabetes Through Inhibition of T-Cell Receptor-Induced NF-κB Signaling. Diabetes 2017, 66, 2033–2043.
- Briknarová, K.; Nasertorabi, F.; Havert, M.L.; Eggleston, E.; Hoyt, D.W.; Li, C.; Olson, A.J.; Vuori, K.; Ely, K.R. The Serine-rich Domain from Crk-associated Substrate (p130cas) Is a Four-helix Bundle *. J. Biol. Chem. 2005, 280, 21908–21914.
- Guerrero, M.S.; Parsons, J.T.; Bouton, A.H. Cas and NEDD9 Contribute to Tumor Progression through Dynamic Regulation of the Cytoskeleton. Genes. Cancer 2012, 3, 371–381.
- Tornillo, G.; Defilippi, P.; Cabodi, S. Cas proteins: Dodgy scaffolding in breast cancer. Breast Cancer Res. 2014, 16, 1–9.
- Heissler, S.M.; Sellers, J.R. Myosins. In Encyclopedia of Cell Biology; Bradshaw, R.A., Stahl, P.D., Eds.; Academic Press: Waltham, MA, USA, 2016; pp. 597–607.
- Rogers, S.L.; Gelfand, V.I. Membrane trafficking, organelle transport, and the cytoskeleton. Curr. Opin. Cell Biol. 2000, 12, 57–62.
- Li, S.; Mecca, A.; Kim, J.; Caprara, G.A.; Wagner, E.L.; Du, T.-T.; Petrov, L.; Xu, W.; Cui, R.; Rebustini, I.T.; et al. Myosin-VIIa is expressed in multiple isoforms and essential for tensioning the hair cell mechanotransduction complex. Nat. Commun. 2020, 11, 2066.
- Ehl, S.; de Saint Basile, G. Chapter 20—Genetic Diseases Predisposing to HLH. In Stiehm’s Immune Deficiencies; Sullivan, K.E., Stiehm, E.R., Eds.; Academic Press: Amsterdam, The Netherlands, 2014; pp. 437–460.
- Navinés-Ferrer, A.; Martín, M. Long-Tailed Unconventional Class I Myosins in Health and Disease. Int. J. Mol. Sci. 2020, 21, 2555.
- Berg, J.S.; Powell, B.C.; Cheney, R.E. A millennial myosin census. Mol. Biol. Cell 2001, 12, 780–794.
- Fili, N.; Toseland, C.P. Unconventional Myosins: How Regulation Meets Function. Int. J. Mol. Sci. 2020, 21, 67.
- Woolner, S.; Bement, W.M. Unconventional myosins acting unconventionally. Trends Cell Biol. 2009, 19, 245–252.
- Kurochkina, N.; Guha, U. SH3 domains: Modules of protein–protein interactions. Biophys. Rev. 2013, 5, 29–39.
- Bi, J.; Chase, S.E.; Pellenz, C.D.; Kurihara, H.; Fanning, A.S.; Krendel, M. Myosin 1e is a component of the glomerular slit diaphragm complex that regulates actin reorganization during cell-cell contact formation in podocytes. Am. J. Physiol. -Ren. Physiol. 2013, 305, F532–F544.
- Yu, I.-M.; Planelles-Herrero, V.J.; Sourigues, Y.; Moussaoui, D.; Sirkia, H.; Kikuti, C.; Stroebel, D.; Titus, M.A.; Houdusse, A. Myosin 7 and its adaptors link cadherins to actin. Nat. Commun. 2017, 8, 15864.
- Grimshaw, S.J.; Mott, H.R.; Stott, K.M.; Nielsen, P.R.; Evetts, K.A.; Hopkins, L.J.; Nietlispach, D.; Owen, D. Structure of the Sterile α Motif (SAM) Domain of the Saccharomyces cerevisiae Mitogen-activated Protein Kinase Pathway-modulating Protein STE50 and Analysis of Its Interaction with the STE11 SAM*. J. Biol. Chem. 2004, 279, 2192–2201.
- Qiao, F.; Bowie, J.U. The Many Faces of SAM. Sci. STKE 2005, 2005, re7.
- Grossmann, A.; Benlasfer, N.; Birth, P.; Hegele, A.; Wachsmuth, F.; Apelt, L.; Stelzl, U. Phospho-tyrosine dependent protein–protein interaction network. Mol. Syst. Biol. 2015, 11, 794.
- Toonen, R.F.G.; Verhage, M. Vesicle trafficking: Pleasure and pain from SM genes. Trends Cell Biol. 2003, 13, 177–186.
- Offenhäuser, N.; Borgonovo, A.; Disanza, A.; Romano, P.; Ponzanelli, I.; Iannolo, G.; Di Fiore, P.P.; Scita, G. The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol. Biol. Cell 2004, 15, 91–98.
- Jaufmann, J.; Franke, F.C.; Sperlich, A.; Blumendeller, C.; Kloos, I.; Schneider, B.; Sasaki, D.; Janssen, K.P.; Beer-Hammer, S. The emerging and diverse roles of the SLy/SASH1-protein family in health and disease-Overview of three multifunctional proteins. Faseb J. 2021, 35, e21470.
- Kwan, J.J.; Slavkovic, S.; Piazza, M.; Wang, D.; Dieckmann, T.; Johnson, P.E.; Wen, X.-Y.; Donaldson, L.W. HACS1 signaling adaptor protein recognizes a motif in the paired immunoglobulin receptor B cytoplasmic domain. Commun. Biol. 2020, 3, 672.
- Bencsik, N.; Pusztai, S.; Borbély, S.; Fekete, A.; Dülk, M.; Kis, V.; Pesti, S.; Vas, V.; Szűcs, A.; Buday, L.; et al. Dendritic spine morphology and memory formation depend on postsynaptic Caskin proteins. Sci. Rep. 2019, 9, 16843.
- Kwan, J.J.; Donaldson, L.W. A lack of peptide binding and decreased thermostability suggests that the CASKIN2 scaffolding protein SH3 domain may be vestigial. BMC Struct. Biol. 2016, 16, 14.
- Yamada, M.; Ishii, N.; Asao, H.; Murata, K.; Kanazawa, C.; Sasaki, H.; Sugamura, K. Signal-transducing adaptor molecules STAM1 and STAM2 are required for T-cell development and survival. Mol. Cell Biol. 2002, 22, 8648–8658.
- Kim, H.; Oh, H.; Oh, Y.S.; Bae, J.; Hong, N.H.; Park, S.J.; Ahn, S.; Lee, M.; Rhee, S.; Lee, S.H.; et al. SPIN90, an adaptor protein, alters the proximity between Rab5 and Gapex5 and facilitates Rab5 activation during EGF endocytosis. Exp. Mol. Med. 2019, 51, 1–14.
- Davis, R.J. Signal transduction by the JNK group of MAP kinases. Cell 2000, 103, 239–252.
- Radhakrishnan, H.; Walther, W.; Zincke, F.; Kobelt, D.; Imbastari, F.; Erdem, M.; Kortüm, B.; Dahlmann, M.; Stein, U. MACC1-the first decade of a key metastasis molecule from gene discovery to clinical translation. Cancer Metastasis Rev. 2018, 37, 805–820.
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13.
- Esmailzadeh, S.; Jiang, X. AHI-1: A novel signaling protein and potential therapeutic target in human leukemia and brain disorders. Oncotarget 2011, 2, 918–934.
- Butt, E.; Howard, C.M.; Raman, D. LASP1 in Cellular Signaling and Gene Expression: More than Just a Cytoskeletal Regulator. Cells 2022, 11, 3817.
- Jung, S.E.; Choi, J.W.; Moon, H.; Oh, S.; Lim, S.; Lee, S.; Kim, S.W.; Hwang, K.C. Small G protein signaling modulator 3 (SGSM3) knockdown attenuates apoptosis and cardiogenic differentiation in rat mesenchymal stem cells exposed to hypoxia. PLoS ONE 2020, 15, e0231272.
- Carrizosa, E. Role and Regulation of the Actin-Regulatory Protein HS1 in TCR Signaling. Ph.D. Thesis, University of Pennsylvania, Philadelphia, PA, USA, 2009.
- Cosen-Binker, L.I.; Kapus, A. Cortactin: The gray eminence of the cytoskeleton. Physiology 2006, 21, 352–361.
- Yuen, M.; Ottenheijm, C.A.C. Nebulin: Big protein with big responsibilities. J. Muscle Res. Cell Motil. 2020, 41, 103–124.
- Krause, M.; Sechi, A.S.; Konradt, M.; Monner, D.A.; Gertler, F.B.; Wehland, J. Fyn-Binding Protein (Fyb)/Slp-76–Associated Protein (Slap), Ena/Vasodilator-Stimulated Phosphoprotein (Vasp) Proteins and the Arp2/3 Complex Link T Cell Receptor (Tcr) Signaling to the Actin Cytoskeleton. J. Cell Biol. 2000, 149, 181–194.
- Arnaud, E.; Zenker, J.; de Preux Charles, A.S.; Stendel, C.; Roos, A.; Médard, J.J.; Tricaud, N.; Kleine, H.; Luscher, B.; Weis, J.; et al. SH3TC2/KIAA1985 protein is required for proper myelination and the integrity of the node of Ranvier in the peripheral nervous system. Proc. Natl. Acad. Sci. USA 2009, 106, 17528–17533.
- Zelazny, E.; Ivanov, R.; Gaude, T. The Plant SNX Family and Its Role in Endocytosis. In Endocytosis in Plants; Šamaj, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 233–247.
- Chen, Y.; He, F.; Wang, R.; Yao, M.; Li, Y.; Guo, D.; He, S. NCF1/2/4 Are Prognostic Biomarkers Related to the Immune Infiltration of Kidney Renal Clear Cell Carcinoma. Biomed. Res. Int. 2021, 2021, 5954036.
- Taylor, J.P.; Tse, H.M. The role of NADPH oxidases in infectious and inflammatory diseases. Redox Biol. 2021, 48, 102159.
- Scott, K.A.; Batey, S.; Hooton, K.A.; Clarke, J. The folding of spectrin domains I: Wild-type domains have the same stability but very different kinetic properties. J. Mol. Biol. 2004, 344, 195–205.
- Machnicka, B.; Czogalla, A.; Hryniewicz-Jankowska, A.; Bogusławska, D.M.; Grochowalska, R.; Heger, E.; Sikorski, A.F. Spectrins: A structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim. Et Biophys. Acta (BBA)—Biomembr. 2014, 1838, 620–634.
- Hale, J.; An, X.; Guo, X.; Gao, E.; Papoin, J.; Blanc, L.; Hillyer, C.D.; Gratzer, W.; Baines, A.; Mohandas, N. αI-spectrin represents evolutionary optimization of spectrin for red blood cell deformability. Biophys. J. 2021, 120, 3588–3599.
- Mosaddeghzadeh, N.; Ahmadian, M.R. The RHO Family GTPases: Mechanisms of Regulation and Signaling. Cells 2021, 10, 1831.
- Kunimura, K.; Uruno, T.; Fukui, Y. DOCK family proteins: Key players in immune surveillance mechanisms. Int. Immunol. 2020, 32, 5–15.
- Hanawa-Suetsugu, K.; Kukimoto-Niino, M.; Mishima-Tsumagari, C.; Akasaka, R.; Ohsawa, N.; Sekine, S.-i.; Ito, T.; Tochio, N.; Koshiba, S.; Kigawa, T.; et al. Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. Proc. Natl. Acad. Sci. USA 2012, 109, 3305–3310.
- Laurin, M.; Côté, J.F. Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes. Dev. 2014, 28, 533–547.
- Gumienny, T.L.; Brugnera, E.; Tosello-Trampont, A.-C.; Kinchen, J.M.; Haney, L.B.; Nishiwaki, K.; Walk, S.F.; Nemergut, M.E.; Macara, I.G.; Francis, R.; et al. CED-12/ELMO, a Novel Member of the CrkII/Dock180/Rac Pathway, Is Required for Phagocytosis and Cell Migration. Cell 2001, 107, 27–41.
- Tu, Y.; Kucik, D.F.; Wu, C. Identification and kinetic analysis of the interaction between Nck-2 and DOCK180. FEBS Lett. 2001, 491, 193–199.
- Côté, J.-F.o.; Vuori, K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 2002, 115, 4901–4913.
- Akakura, S.; Kar, B.; Singh, S.; Cho, L.; Tibrewal, N.; Sanokawa-Akakura, R.; Reichman, C.; Ravichandran, K.S.; Birge, R.B. C-terminal SH3 domain of CrkII regulates the assembly and function of the DOCK180/ELMO Rac-GEF. J. Cell. Physiol. 2005, 204, 344–351.
- Griffiths, E.K.; Krawczyk, C.; Kong, Y.-Y.; Raab, M.; Hyduk, S.J.; Bouchard, D.; Chan, V.S.; Kozieradzki, I.; Oliveira-dos-Santos, A.J.; Wakeham, A.; et al. Positive Regulation of T Cell Activation and Integrin Adhesion by the Adapter Fyb/Slap. Science 2001, 293, 2260–2263.
- Bauer, R.; Humphries, M.; Fässler, R.; Winklmeier, A.; Craig, S.E.; Bosserhoff, A.-K. Regulation of integrin activity by MIA. J. Biol. Chem. 2006, 281, 11669–11677.
- Sasahira, T.; Kirita, T.; Nishiguchi, Y.; Kurihara, M.; Nakashima, C.; Bosserhoff, A.K.; Kuniyasu, H. A comprehensive expression analysis of the MIA gene family in malignancies: MIA gene family members are novel, useful markers of esophageal, lung, and cervical squamous cell carcinoma. Oncotarget 2016, 7, 31137–31152.
- Winklmeier, A.; Contreras-Shannon, V.; Arndt, S.; Melle, C.; Bosserhoff, A.K. Cadherin-7 interacts with melanoma inhibitory activity protein and negatively modulates melanoma cell migration. Cancer Sci. 2009, 100, 261–268.
- Feng, Z.; Yang, K.; Pastor-Pareja, J.C. Tales of the ER-Golgi Frontier: Drosophila-Centric Considerations on Tango1 Function. Front. Cell Dev. Biol. 2020, 8, 619022.
- Rendtorff, N.D.; Frödin, M.; Attié-Bitach, T.; Vekemans, M.; Tommerup, N. Identification and Characterization of an Inner Ear-Expressed Human Melanoma Inhibitory Activity (MIA)-like Gene (MIAL) with a Frequent Polymorphism That Abolishes Translation. Genomics 2001, 71, 40–52.
- McConnachie, D.J.; Stow, J.L.; Mallett, A.J. Ciliopathies and the Kidney: A Review. Am. J. Kidney Dis. 2021, 77, 410–419.
- Moog-Lutz, C.; Peterson, E.J.; Lutz, P.G.; Eliason, S.; Cavé-Riant, F.; Singer, A.; Di Gioia, Y.; Dmowski, S.; Kamens, J.; Cayre, Y.E. PRAM-1 is a novel adaptor protein regulated by retinoic acid (RA) and promyelocytic leukemia (PML)-RA receptor α in acute promyelocytic leukemia cells. J. Biol. Chem. 2001, 276, 22375–22381.
This entry is offline, you can click here to edit this entry!
