Cyanobacteria are naturally capable of producing valuable secondary metabolites through photosynthesis. These photosynthetic bacteria can be further improved via engineering to produce more in terms of diversity and yield. Recent advances in systems and synthetic biology approaches are being adopted in the cyanobacterial engineering field to push the industrial capabilities even further.
- cyanobacteria
- photosynthesis
- secondary metabolites
- metabolic engineering
- synthetic biology
- systems biology
- genome-scale model
1. Introduction
Cyanobacteria are oxygenic photosynthetic bacteria that can produce various secondary metabolites. Given the ability to utilize sunlight and atmospheric carbon dioxide (CO2) as a part of the renewable photosynthetic process, cyanobacteria are considered sustainable bioproduction hosts [1]. A number of secondary metabolites naturally synthesized by cyanobacteria, such as carotenoids, phycocyanins, and squalene, are used in the pharmaceutical, cosmetic, and healthcare industries [2][3][4]. In addition, owing to their rapid growth and increased scope for engineering, multiple efforts have been made to utilize cyanobacteria as production hosts for valuable biochemicals by introducing heterologous pathways [5][6].
While continuous development has been reported in metabolic engineering strategies for producing biochemicals in bacterial hosts, the synthetic biology approach accelerated the development by providing diverse genetic parts and engineering tools. For other model platforms such as Escherichia coli, there is an abundant catalog of genetic parts including synthetic promoters and ribosome binding sites (RBSs), which have been successfully introduced to improve gene expression in heterologous pathways [7]. However, owing to the lack of genetic parts for pathway engineering in cyanobacteria, application of metabolic engineering tools is limited [8]. Thus, development of various tools for pathway engineering and subsequent engineering strategies are required for industrial-scale production of target compounds in cyanobacteria.
With the recent progress in systems biology, genome-wide information of diverse layers such as the genome, transcriptome, translatome, proteome, metabolome, and interactome are being constantly accumulated [9]. Massive amounts of data formed the basis for establishment and development of an in silico genome-scale model (GEM) [10]. It is expected that the application of system-level approaches with the integration of omics data and GEM would address the existing limitations of cyanobacterial engineering.
2. Secondary Metabolite Production by Cyanobacteria
Bacteria produce two kinds of metabolites: primary metabolites essential for survival and secondary metabolites required for auxiliary purposes, such as stress responses, defense mechanisms, metal carrying, and signaling [11]. Secondary metabolites include terpenes, alkaloids, polyketides (PKs), non-ribosomal peptides (NRPs), and ribosomally synthesized and post-translationally modified peptides (RiPPs), which are produced via biosynthetic gene clusters (BGCs). BGCs are clusters of genes positioned in approximate proximity to each other for the production and processing of a compound. Cyanobacteria, being rich in BGCs, are capable of producing diverse secondary metabolites for various purposes, including toxins for defenses or protectants for relieving photodamage and oxidative stress (Table 1).
Table 1. Bioactive secondary metabolites produced in cyanobacteria.
|
Class |
Metabolite |
Bioactivity |
Producing Species |
Ref. |
|
Terpene |
Phycocyanin |
Antioxidant, anti-inflammatory, neuroprotective, hepatoprotective |
All cyanobacteria |
|
|
Terpene |
Carotenoids |
Antioxidant, sunscreen |
All cyanobacteria |
|
|
Terpene |
Squalene |
Antioxidant |
Phormidium |
[19] |
|
Alkaloid |
Saxitoxin |
Neurotoxin |
Anabaena, Aphanizomenon, Cylindrospermopsis, Lyngbya, Planktothrix, |
|
|
Indole |
Nostodione |
Antifungal |
Nostoc |
[23] |
|
Indole alkaloid |
Scytonemin |
Anti-inflammatory, sunscreen |
Scytonema, Nostoc |
|
|
Indole alkaloid |
Hapalindole |
Antibacterial, anti-tuberculosis, anticancer |
Hapalosiphon |
|
|
Alkaloid/Polyketide synthase (PKS) |
Anatoxin-a |
Neurotoxin, anti-inflammatory |
Anabaena, Aphanizomenon, Cylindrospermum, Oscillatoria, Planktothrix |
|
|
Alkaloid/PKS |
Aplysiatoxin |
Cytotoxin, antiviral |
Moorea |
|
|
Alkaloid/Non-ribosomal peptide synthetase (NRPS) |
Lyngbyatoxin |
Cytotoxin, dermatotoxin |
Moorea |
[34] |
|
Alkaloid/PKS-NRPS |
Cylindrospermopsin |
Cytotoxin |
Aphanizomenon, Cylindrospermopsis, Oscillatoria, Raphidiopsis |
|
|
PKS |
Fischerellin |
Antifungal, antialgal, anti-cyanobacterial |
Fischerella |
[38] |
|
NRPS |
β-N-methylamino-l-alanine |
Neurotoxin |
Anabaena, Nostoc |
[39] |
|
NRPS |
Cyanopeptolin |
Protease inhibitor |
Planktothrix, Microcystis |
|
|
PKS-NRPS |
Microcystin |
Hepatotoxin |
Microcystis, Nostoc, Planktothrix, Anabaena |
|
|
PKS-NRPS |
Nodularin |
Hepatotoxin |
Nodularia |
[46] |
|
PKS-NRPS |
Apratoxin |
Anticancer |
Lyngbya |
[47] |
|
PKS-NRPS |
Aeruginoside |
Protease inhibitor |
Planktothrix |
[48] |
|
PKS-NRPS |
Aeruginosin |
Protease inhibitor |
Microcystis, Planktothrix |
|
|
PKS-NRPS |
Cryptophycins |
Cytotoxin |
Nostoc |
[50] |
|
PKS-NRPS |
Nostophycins |
Cytotoxin |
Nostoc |
[51] |
|
PKS-NRPS |
Curacins |
Cytotoxin |
Moorea |
[52] |
|
PKS-NRPS |
Hectochlorin |
Cytotoxin |
Moorea |
[53] |
|
PKS-NRPS |
Jamaicamides |
Neurotoxin |
Moorea |
[54] |
|
PKS-NRPS |
Dolastatin |
Cytotoxin, anticancer, antiprotozoal |
Moorea, Lyngbya, Symploca |
|
|
Lipopeptide |
Antillatoxin |
Neurotoxin |
Moorea |
[57] |
|
Lipopeptide |
Carmabin |
Antimalarial, anticancer, antiproliferative |
Moorea |
|
|
Lipopeptide |
Lyngbyabellin |
Cytotoxin, antifungal |
Moorea, Lyngbya |
|
|
Lipopeptide |
Kalkitoxin |
Neurotoxin |
Moorea |
[57] |
|
Ribosomally synthesized and post-translationally modified peptide (RiPP) |
Patellamide |
Moderate cytotoxicity |
Prochloron |
[62] |
|
RiPP |
Microviridin |
Protease inhibitor |
Microcystis, Planktothrix |
|
|
RiPP |
Shinorin |
Sunscreen |
Anabaena, Nostoc |
[65] |
|
Fatty acid amide |
Besarhanamide A |
Moderate toxicity to brine shrimp |
Moorea |
[66] |
|
Fatty acid amide |
Semiplenamide |
Toxicity to brine shrimp |
Lyngbya |
[67] |
|
Lipopolysaccharide |
Lipopolysaccharides |
Endotoxin |
All cyanobacteria |
[68] |
|
Polysaccharide |
Polysaccharide |
Antitumor, antiviral, antibacterial, anti-inflammatory, immunostimulant |
All cyanobacteria |
|
|
Nucleoside |
Toyocamycin |
Antifungal |
Tolypothrix |
[72] |
|
Nucleoside |
Tubercidin |
Antifungal |
Tolypothrix |
[73] |
2.1. Prediction of Biosynthetic Gene Clusters (BGCs) in Cyanobacterial Genomes
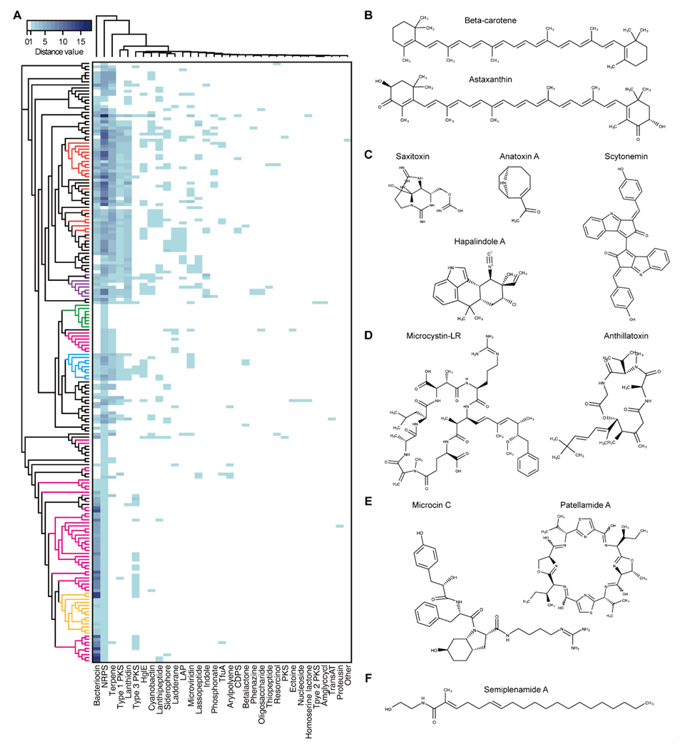
To investigate the secondary metabolites produced by cyanobacteria, 196 complete genome sequences of cyanobacteria available at the National Center for Biotechnology Information (NCBI) genome portal were inspected for BGCs using antiSMASH [74]. Thirty-three different types of BGCs were identified. The 196 complete genome sequences of cyanobacteria used in the BGC search were arranged according to the phylogenetic tree. The heatmap representing the numbers of each type of BGC found in each cyanobacterium showed that the cyanobacteria from the same genera had similar classes and numbers of BGCs (Figure 1A). It was evident that a single genome contained several BGCs with multiple occurrences. In particular, there were cyanobacteria with large number of bacteriocin, terpene, and non-ribosomal peptide synthetase (NRPS) BGCs, which accounted for 74.4% of the total predicted BGCs (n = 2119). For example, it was predicted that the genome of Moorea producens PAL-8-15-08-1 carries 18 NRPS BGCs. The most widely distributed BGC was the terpene BGC, which was found in all cyanobacteria except for two species (Limnospira fusiformis SAG 85.79 and Nodularia spumigena UHCC 0039). Terpene is essential for photosynthetic organisms. Undetected terpene BGCs in the two species could have resulted from the deviations in the BGC search criteria of antiSMASH. The 33 BGCs were classified according to their structural and functional similarities to the following categories: terpene, indole, PK synthase (PKS)/NRPS (type 1, 2, 3 PKSs, NRPS, cyclodipeptide synthase-based tRNA-dependent peptide, resorcinol, and siderophore), RiPP (bacteriocin, lanthidin, linear azole-containing peptide, microviridin, lasso peptide, cyanobactin, thiopeptide, trifolitoxin, proteusin, and lanthipeptide), lipid/saccharide/nucleoside (heterocyst glycolipid synthase, ladderane, arylpolyene, aminoglycoside/aminocyclitol, oligosaccharide, and nucleoside), and others (phosphonate, phenazine, ectoine, β-lactone, and homoserine lactone).
2.2. Terpenes
Terpene is a family of compounds with varying structures that occupies a large proportion of the natural products [75]. Terpenes are mainly produced by plants or fungi, as well as the bacterial species via mevalonate (MVA) pathway or methylerythritol-phosphate (MEP) pathway using acetyl-CoA or glyceraldehyde 3-phosphate and pyruvate as substrates [76]. While MVA and MEP pathways are mutually exclusive in most organisms, cyanobacteria mainly utilize the MEP pathway, using substrates generated during photosynthesis. The MEP pathway produces isomeric 5-carbon compounds, isopentyl pyrophosphate (IPP), and dimethylallyl pyrophosphate (DMAPP), which are further condensed into geranyl pyrophosphate (GPP), the building block in terpene biosynthesis. From the GPP, terpenes of varying structures can be generated. Terpenes conduct various cellular processes necessary for survival, such as the ubiquinone in the electron transport chain associated with cellular respiration, chlorophyll, carotenoids, and plastoquinones in photosynthetic processes, and hopanoids in cell membrane biosynthesis and stability (Figure 1B) [77]. In particular, photosynthetic cyanobacteria contain a wide variety of carotenoids. Most of the genome-sequenced cyanobacteria have β-carotene BGC. Production of other carotenoids, such as zeaxanthin and nostoxanthin are dependent on the presence of carotenogenesis pathway connected to β-carotene [3][78]. The terpene compounds, including the carotenoids obtained from cyanobacteria are of industrial value owing to their various applications. For example, β-carotene, astaxanthin, and canthaxanthin are used as color additives or animal feeds. Phycocyanin exhibits anti-oxidant, anti-inflammatory, neuroprotective, and hepatoprotective effects [2][13][79].
Figure 1. Cyanobacterial secondary metabolites. (A) Heatmap of the predicted cyanobacterial secondary metabolite biosynthstic gene clusters (BGCs). The left-most phylogenetic tree is constructed by up-to-date bacterial core gene (UBCG) phylogenetic analysis of the 196 cyanobacterial complete genome sequences. The evolutionary distances were provided by UBCG and plotted by RAxML [80][81]. The tree is not to scale. Red: Nostoc, purple: Calothrix, green: Synechocystis, pink: Synechoccus, blue: Microcystis, and yellow: Prochlorococcus. (B–F) Molecular structures of cyanobacterial secondary metabolites. (B) Terpenes, (C) alkaloids, (D) polyketides (PKs), non-ribosomal peptides (NRPs), (E) RiPPs, and (F) fatty acid amide. Abbreviations; NRPS, non-ribosomal peptide synthetase; HglE, heterocyst glycolipid synthase; LAP, linear azol(in)e-containing peptide; TfuA, ribosomally synthesized peptide antibiotic trifolitoxin; CDPS, cyclodipeptide synthase-based tRNA dependent peptide; PKS, polyketide synthase; Amglyccycl, aminoglycosides/aminocyclitols; TransAT, trans-acyltransferase type I PKS.
2.3. Alkaloids
Alkaloids comprise various nitrogen containing compounds that are produced from diverse organisms, including fungi, plants, bacteria, and animals. Alkaloids produced by cyanobacteria often show toxic characteristics. For example, the anatoxin-a produced by species of the Anabaena genera is a neurotoxin that binds irreversibly to nicotinic acetylcholine receptors causing paralysis or even death in fish and mammals (Figure 1C) [31]. Anatoxin-a is also categorized as a PK, which is synthesized by PKS [82]. Another well-known example, saxitoxin, blocks the sodium (Na+) channels in shellfish and induces paralytic shellfish poisoning in humans on consumption of saxitoxin-accumulated seafood. The chemical derivatives carrying the indole rings are classified as indole alkaloids. They are biosynthesized using tryptophan as a precursor. Cyanobacterial indole alkaloids have diverse functions. For example, the hapalindole synthesized from cyanobacteria Hapalosiphon fontinalis exhibits antibacterial, anti-tuberculosis, and anticancer activities [83]. In addition, the scytonemin produced by Scytonema sp. renders photoprotective effects to the cyanobacterial cells by absorbing the harmful ultraviolet (UV)-A radiation [84].
2.4. Polyketides/Non-Ribosomal Peptide/Lipopeptides/Siderophores
PKS and NRPS are representatives of enzymes responsible for the biosynthesis of secondary metabolites in various organisms. Enzymes of these classes consists of at least three essential modular domains that facilitate chain elongation and modification [85]. First, the catalytic domain binds to and activates the building block, which then is transferred to the carrier protein domain. Second, the carrier protein domain loads the activated building block to the growing PK/NRP chain it holds. Third, the other catalytic domain catalyzes the bond formation between the growing chain and the newly loaded building block. PKS and NRPS differ in their use of precursors for the building block. While PKS utilizes malonyl-CoA or methylmalonyl-CoA, the NRPS uses proteinogenic and non-proteinogenic amino acid monomers. In addition, there are cases wherein compounds are synthesized via the PKS–NRPS hybrid system. A well-known example could be microcystin, the BGC of which contains two PKS, single PKS–NRPS, and three NRPS [42][86]. Microcystin produced from various cyanobacterial species belonging to the genus Microcystis, Nostoc, Planktothrix, and Anabaena, shows hepatotoxic activity in humans (Figure 1D). Various other toxins synthesized by the PKS, NRPS, or PKS–NRPS hybrid system includes lyngbyatoxin, apratoxin, and aplysiatoxin.
The NRPS includes lipopeptides owing to their lipid linked peptide structures synthesized by a combination of lipid tails and amino acids. Examples of lipopeptides include antillatoxin and carmabin from M. producens, and lyngbyabellin from M. bouillonii (Figure 1D). Antillatoxin and lyngbyabellin show neurotoxic activity and cytotoxicity, and carmabin exhibit anti-malarial activity. Siderophores are included in the NRPS-produced compounds. Iron is essential for bacterial survival. However, since it exists in an insoluble form in the environment, some bacteria have evolved to facilitate iron uptake by producing small molecules with high affinity to ferric iron, called siderophores.
2.5. Ribosomally Synthesized and Post-Translationally Modified Peptides
RiPP is a class of secondary metabolites that includes, as its name depicts, ribosomally synthesized and post-translationally modified peptides. Post-translational modifications include leader peptide hydrolysis, cyclization, and disulfide bond formation. RiPP BGC generally consists of a short precursor peptide with an N-terminal leader and a C-terminal core sequence, and post-translational modification (PTM) enzymes [87][88]. The PTM enzymes shape the linear peptide by several modifications that provide structural and functional diversity to the mature scaffold. Compounds that were previously classified as lanthipeptide, lasso peptide, microviridin, cyanobactin, and microcin are now re-classified under RiPP, which have a broad range of bioactivities such as protease inhibition, cytotoxicity, signaling, anti-cancer, and anti-human immunodeficiency virus (anti-HIV) (Figure 1E) [87]. For example, microviridin, which was first isolated from M. viridis, is a serine protease inhibitor, and patellamide A produced by Prochloron didemni has moderate cytotoxicity [62].
2.6. Lipids/Saccharides/Nucleosides/Others
Lipids, saccharides, and nucleosides are generally categorized as primary metabolites. However, there are exceptions, when they are considered as secondary metabolites instead of primary metabolites. For example, besarhanamide A and semiplenamide exhibiting toxicity against brine shrimp are fatty acid amides isolated from M. producens and Lyngbya semiplena, respectively (Figure 1F) [66][89]. It is known that cyanobacterium Cyanothece sp. 113 can produce up to 22 g/L of polysaccharide, which exceeds the producing ability of eukaryotic microalgae, such as Dunaliella salina [90][91]. Polysaccharides are generally used as stabilization or thickening agents for emulsions. In some cases, they are used as bioactive compounds owing to their antitumor, antiviral, antibacterial, anti-inflammatory, and immunostimulatory properties [92][93][94][95]. Toyocamycin and tubercidin are both anti-fungal nucleoside chemicals isolated from Tolypothrix tenuis [96]. In addition, a small number of phosphonate, phenazine, ectoine, and β-lactone BGC were also detected.
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms8121849
References
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A precious bio-resource in agriculture, ecosystem, and environmental sustainability. Microbiol. 2016, 7, 529.
- Mogany, T.; Swalaha, F.M.; Kumari, S.; Bux, F. Elucidating the role of nutrients in C-phycocyanin production by the halophilic cyanobacterium Euhalothece J. Appl. Phycol. 2018, 30, 2259–2271.
- Liang, C.; Zhao, F.; Wei, W.; Wen, Z.; Qin, S. Carotenoid biosynthesis in cyanobacteria: Structural and evolutionary scenarios based on comparative genomics. J. Biol. Sci. 2006, 2, 197.
- Lee, H.J.; Lee, J.; Lee, S.-M.; Um, Y.; Kim, Y.; Sim, S.J.; Choi, J.-I.; Woo, H.M. Direct conversion of CO2 to α-farnesene using metabolically engineered Synechococcus elongatus PCC 7942. Agric. Food Chem. 2017, 65, 10424–10428.
- Kanno, M.; Carroll, A.L.; Atsumi, S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Commun. 2017, 8, 1–11.
- Gao, X.; Gao, F.; Liu, D.; Zhang, H.; Nie, X.; Yang, C. Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 2016, 9, 1400–1411.
- Stephanopoulos, G. Synthetic biology and metabolic engineering. ACS Synth. Biol. 2012, 1, 514–525.
- Ramey, C.J.; Barón-Sola, A.n.; Aucoin, H.R.; Boyle, N.R. Genome engineering in cyanobacteria: Where we are and where we need to go. ACS Synth. Biol. 2015, 4, 1186–1196.
- Lin, W.-R.; Tan, S.-I.; Hsiang, C.-C.; Sung, P.-K.; Ng, I.-S. Challenges and opportunity of recent genome editing and multi-omics in cyanobacteria and microalgae for biorefinery. Technol. 2019, 291, 121932.
- Broddrick, J.T.; Rubin, B.E.; Welkie, D.G.; Du, N.; Mih, N.; Diamond, S.; Lee, J.J.; Golden, S.S.; Palsson, B.O. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Natl. Acad. Sci. USA 2016, 113, E8344–E8353.
- Kultschar, B.; Llewellyn, C. Secondary metabolites in cyanobacteria. In Secondary Metabolites—Sources and Applications; IntechOpen: London, UK, 2018; pp. 23–36.
- Romay, C.; Armesto, J.; Remirez, D.; Gonzalez, R.; Ledon, N.; Garcia, I. Antioxidant and anti-inflammatory properties of C-phycocyanin from blue-green algae. Res. 1998, 47, 36–41, doi:10.1007/s000110050256.
- Romay, C.; Gonzalez, R.; Ledon, N.; Remirez, D.; Rimbau, V. C-phycocyanin: A biliprotein with antioxidant, anti-inflammatory and neuroprotective effects. Protein Pept. Sci. 2003, 4, 207–216, doi:10.2174/1389203033487216.
- Benedetti, S.; Benvenuti, F.; Pagliarani, S.; Francogli, S.; Scoglio, S.; Canestrari, F. Antioxidant properties of a novel phycocyanin extract from the blue-green alga Aphanizomenon flos-aquae. Life Sci. 2004, 75, 2353–2362, doi:10.1016/j.lfs.2004.06.004.
- Kuddus, M.; Singh, P.; Thomas, G.; Al-Hazimi, A. Recent developments in production and biotechnological applications of C-phycocyanin. Biomed Res. Int. 2013, 2013, 742859, doi:10.1155/2013/742859.
- Patel, A.; Mishra, S.; Ghosh, P.K. Antioxidant potential of C-phycocyanin isolated from cyanobacterial species Lyngbya, Phormidium and Spirulina Indian J. Biochem. Biophys. 2006, 43, 25–31.
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Asp. Med. 2003, 24, 345–351, doi:10.1016/s0098-2997(03)00030-x.
- Wada, N.; Sakamoto, T.; Matsugo, S. Multiple roles of photosynthetic and sunscreen pigments in cyanobacteria focusing on the oxidative stress. Metabolites 2013, 3, 463–483, doi:10.3390/metabo3020463.
- Fagundes, M.B.; Falk, R.B.; Facchi, M.M.X.; Vendruscolo, R.G.; Maroneze, M.M.; Zepka, L.Q.; Jacob-Lopes, E.; Wagner, R. Insights in cyanobacteria lipidomics: A sterols characterization from Phormidium autumnale biomass in heterotrophic cultivation. Food Res. Int. 2019, 119, 777–784, doi:10.1016/j.foodres.2018.10.060.
- Kellmann, R.; Mihali, T.K.; Neilan, B.A. Identification of a saxitoxin biosynthesis gene with a history of frequent horizontal gene transfers. Mol. Evol. 2008, 67, 526–538, doi:10.1007/s00239-008-9169-2.
- Mihali, T.K.; Kellmann, R.; Neilan, B.A. Characterisation of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon NH-5. BMC Biochem. 2009, 10, 8, doi:10.1186/1471-2091-10-8.
- Murray, S.A.; Wiese, M.; Stuken, A.; Brett, S.; Kellmann, R.; Hallegraeff, G.; Neilan, B.A. sxtA-based quantitative molecular assay to identify saxitoxin-producing harmful algal blooms in marine waters. Environ. Microbiol. 2011, 77, 7050–7057, doi:10.1128/AEM.05308-11.
- Burja, A.M.; Banaigs, B.; Abou-Mansour, E.; Burgess, J.G.; Wright, P.C.J.T. Marine cyanobacteria—A prolific source of natural products. Tetrahedron 2001, 57, 9347–9377.
- Rastogi, R.P.; Sonani, R.R.; Madamwar, D. Cyanobacterial sunscreen scytonemin: Role in photoprotection and biomedical research. Biochem. Biotechnol. 2015, 176, 1551–1563, doi:10.1007/s12010-015-1676-1.
- Garcia‐Pichel, F.; Castenholz, R.W.J.J.O.P. Characterization and biological implications of scytonemin, a cyanobacterial sheath pigment. Phycol. 1991, 27, 395–409.
- Proteau, P.J.; Gerwick, W.H.; Garcia-Pichel, F.; Castenholz, R. The structure of scytonemin, an ultraviolet sunscreen pigment from the sheaths of cyanobacteria. Experientia 1993, 49, 825–829, doi:10.1007/BF01923559.
- Soule, T.; Stout, V.; Swingley, W.D.; Meeks, J.C.; Garcia-Pichel, F. Molecular genetics and genomic analysis of scytonemin biosynthesis in Nostoc punctiforme ATCC 29133. Bacteriol. 2007, 189, 4465–4472, doi:10.1128/JB.01816-06.
- Klein, D.; Daloze, D.; Braekman, J.C.; Hoffmann, L.; Demoulin, V. New hapalindoles from the cyanophyte Hapalosiphon laingii. Nat. Prod. 1995, 58, 1781–1785.
- Moore, R.E.; Cheuk, C.; Patterson, G.M.L. Hapalindoles: New alkaloids from the blue-green alga Hapalosiphon fontinalis. Am. Chem. Soc. 1984, 106, 6456–6457.
- Mejean, A.; Mann, S.; Maldiney, T.; Vassiliadis, G.; Lequin, O.; Ploux, O. Evidence that biosynthesis of the neurotoxic alkaloids anatoxin-a and homoanatoxin-a in the cyanobacterium Oscillatoria PCC 6506 occurs on a modular polyketide synthase initiated by L-proline. Am. Chem. Soc. 2009, 131, 7512–7513, doi:10.1021/ja9024353.
- Rantala-Ylinen, A.; Kana, S.; Wang, H.; Rouhiainen, L.; Wahlsten, M.; Rizzi, E.; Berg, K.; Gugger, M.; Sivonen, K. Anatoxin-a synthetase gene cluster of the cyanobacterium Anabaena strain 37 and molecular methods to detect potential producers. Appl. Environ. Microbiol. 2011, 77, 7271–7278, doi:10.1128/AEM.06022-11.
- Moore, R.E.; Blackman, A.J.; Cheuk, C.E.; Mynderse, J.S.; Matsumoto, G.K.; Clardy, J.; Woodard, R.W.; Craig, J.C. Absolute stereochemistries of the aplysiatoxins and oscillatoxin A. Org. Chem. 1984, 49, 2484–2489.
- Gupta, D.K.; Kaur, P.; Leong, S.T.; Tan, L.T.; Prinsep, M.R.; Chu, J.J. Anti-Chikungunya viral activities of aplysiatoxin-related compounds from the marine cyanobacterium Trichodesmium erythraeum. Drugs 2014, 12, 115–127, doi:10.3390/md12010115.
- Edwards, D.J.; Gerwick, W.H. Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. Am. Chem. Soc. 2004, 126, 11432–11433, doi:10.1021/ja047876g.
- Mihali, T.K.; Kellmann, R.; Muenchhoff, J.; Barrow, K.D.; Neilan, B.A. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Environ. Microbiol. 2008, 74, 716–722, doi:10.1128/AEM.01988-07.
- Stuken, A.; Jakobsen, K.S. The cylindrospermopsin gene cluster of Aphanizomenon strain 10E6: Organization and recombination. Microbiology (Reading) 2010, 156, 2438–2451, doi:10.1099/mic.0.036988-0.
- Mazmouz, R.; Chapuis-Hugon, F.; Mann, S.; Pichon, V.; Mejean, A.; Ploux, O. Biosynthesis of cylindrospermopsin and 7-epicylindrospermopsin in Oscillatoria strain PCC 6506: Identification of the cyr gene cluster and toxin analysis. Appl. Environ. Microbiol. 2010, 76, 4943–4949, doi:10.1128/AEM.00717-10.
- Gross, E.M.; Wolk, C.P.; Jüttner, F. Fischerellin, a new allelochemical from the freshwater cyanobacterium Fischerella Muscicola. Phycol. 1991, 27, 686–692.
- Cox, P.A.; Banack, S.A.; Murch, S.J.; Rasmussen, U.; Tien, G.; Bidigare, R.R.; Metcalf, J.S.; Morrison, L.F.; Codd, G.A.; Bergman, B. Diverse taxa of cyanobacteria produce β-N-methylamino-L-alanine, a neurotoxic amino acid. Natl. Acad. Sci. USA 2005, 102, 5074–5078.
- Rounge, T.B.; Rohrlack, T.; Nederbragt, A.J.; Kristensen, T.; Jakobsen, K.S. A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens BMC Genom. 2009, 10, 396, doi:10.1186/1471-2164-10-396.
- Tooming-Klunderud, A.; Rohrlack, T.; Shalchian-Tabrizi, K.; Kristensen, T.; Jakobsen, K.S. Structural analysis of a non-ribosomal halogenated cyclic peptide and its putative operon from Microcystis: Implications for evolution of cyanopeptolins. Microbiology (Reading) 2007, 153, 1382–1393, doi:10.1099/mic.0.2006/001123-0.
- Tillett, D.; Dittmann, E.; Erhard, M.; von Dohren, H.; Borner, T.; Neilan, B.A. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: An integrated peptide-polyketide synthetase system. Biol. 2000, 7, 753–764, doi:10.1016/s1074-5521(00)00021-1.
- Kaneko, T.; Nakajima, N.; Okamoto, S.; Suzuki, I.; Tanabe, Y.; Tamaoki, M.; Nakamura, Y.; Kasai, F.; Watanabe, A.; Kawashima, K.; et al. Complete genomic structure of the bloom-forming toxic cyanobacterium Microcystis aeruginosa NIES-843. DNA Res. 2007, 14, 247–256, doi:10.1093/dnares/dsm026.
- Rouhiainen, L.; Vakkilainen, T.; Siemer, B.L.; Buikema, W.; Haselkorn, R.; Sivonen, K. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Environ. Microbiol. 2004, 70, 686–692, doi:10.1128/aem.70.2.686-692.2004.
- Christiansen, G.; Fastner, J.; Erhard, M.; Borner, T.; Dittmann, E. Microcystin biosynthesis in planktothrix: Genes, evolution, and manipulation. Bacteriol. 2003, 185, 564–572, doi:10.1128/jb.185.2.564-572.2003.
- Moffitt, M.C.; Neilan, B.A. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Environ. Microbiol. 2004, 70, 6353–6362, doi:10.1128/AEM.70.11.6353-6362.2004.
- Grindberg, R.V.; Ishoey, T.; Brinza, D.; Esquenazi, E.; Coates, R.C.; Liu, W.T.; Gerwick, L.; Dorrestein, P.C.; Pevzner, P.; Lasken, R.; et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS ONE 2011, 6, e18565, doi:10.1371/journal.pone.0018565.
- Ishida, K.; Christiansen, G.; Yoshida, W.Y.; Kurmayer, R.; Welker, M.; Valls, N.; Bonjoch, J.; Hertweck, C.; Borner, T.; Hemscheidt, T.; et al. Biosynthesis and structure of aeruginoside 126A and 126B, cyanobacterial peptide glycosides bearing a 2-carboxy-6-hydroxyoctahydroindole moiety. Biol. 2007, 14, 565–576, doi:10.1016/j.chembiol.2007.04.006.
- Ishida, K.; Welker, M.; Christiansen, G.; Cadel-Six, S.; Bouchier, C.; Dittmann, E.; Hertweck, C.; Tandeau de Marsac, N. Plasticity and evolution of aeruginosin biosynthesis in cyanobacteria. Environ. Microbiol. 2009, 75, 2017–2026, doi:10.1128/AEM.02258-08.
- Magarvey, N.A.; Beck, Z.Q.; Golakoti, T.; Ding, Y.; Huber, U.; Hemscheidt, T.K.; Abelson, D.; Moore, R.E.; Sherman, D.H. Biosynthetic characterization and chemoenzymatic assembly of the cryptophycins. Potent anticancer agents from cyanobionts. ACS Chem. Biol. 2006, 1, 766–779, doi:10.1021/cb6004307.
- Fewer, D.P.; Osterholm, J.; Rouhiainen, L.; Jokela, J.; Wahlsten, M.; Sivonen, K. Nostophycin biosynthesis is directed by a hybrid polyketide synthase-nonribosomal peptide synthetase in the toxic cyanobacterium Nostoc strain 152. Appl. Environ. Microbiol. 2011, 77, 8034–8040, doi:10.1128/AEM.05993-11.
- Chang, Z.; Sitachitta, N.; Rossi, J.V.; Roberts, M.A.; Flatt, P.M.; Jia, J.; Sherman, D.H.; Gerwick, W.H. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. Nat. Prod. 2004, 67, 1356–1367, doi:10.1021/np0499261.
- Ramaswamy, A.V.; Sorrels, C.M.; Gerwick, W.H. Cloning and biochemical characterization of the hectochlorin biosynthetic gene cluster from the marine cyanobacterium Lyngbya majuscula. Nat. Prod. 2007, 70, 1977–1986, doi:10.1021/np0704250.
- Edwards, D.J.; Marquez, B.L.; Nogle, L.M.; McPhail, K.; Goeger, D.E.; Roberts, M.A.; Gerwick, W.H. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Biol. 2004, 11, 817–833, doi:10.1016/j.chembiol.2004.03.030.
- Nogle, L.M.; Williamson, R.T.; Gerwick, W.H. Somamides A and B, two new depsipeptide analogues of dolastatin 13 from a Fijian cyanobacterial assemblage of Lyngbya majuscula and Schizothrix J. Nat. Prod. 2001, 64, 716–719, doi:10.1021/np000634j.
- Nogle, L.M.; Gerwick, W.H. Isolation of four new cyclic depsipeptides, antanapeptins A-D, and dolastatin 16 from a Madagascan collection of Lyngbya majuscula. Nat. Prod. 2002, 65, 21–24, doi:10.1021/np010348n.
- Berman, F.W.; Gerwick, W.H.; Murray, T.F. Antillatoxin and kalkitoxin, ichthyotoxins from the tropical cyanobacterium Lyngbya majuscula, induce distinct temporal patterns of NMDA receptor-mediated neurotoxicity. Toxicon 1999, 37, 1645–1648, doi:10.1016/s0041-0101(99)00108-7.
- McPhail, K.L.; Correa, J.; Linington, R.G.; Gonzalez, J.; Ortega-Barria, E.; Capson, T.L.; Gerwick, W.H. Antimalarial linear lipopeptides from a Panamanian strain of the marine cyanobacterium Lyngbya majuscula. Nat. Prod. 2007, 70, 984–988, doi:10.1021/np0700772.
- Hooper, G.J.; Orjala, J.; Schatzman, R.C.; Gerwick, W.H. Carmabins A and B, new lipopeptides from the Caribbean cyanobacterium Lyngbya majuscula. Nat. Prod. 1998, 61, 529–533, doi:10.1021/np970443p.
- Choi, H.; Mevers, E.; Byrum, T.; Valeriote, F.A.; Gerwick, W.H. Lyngbyabellins K-N from two palmyra atoll collections of the marine cyanobacterium Moorea bouillonii. J. Org. Chem. 2012, 2012, 5141–5150, doi:10.1002/ejoc.201200691.
- Han, B.; McPhail, K.L.; Gross, H.; Goeger, D.E.; Mooberry, S.L.; Gerwick, W.H.J.T. Isolation and structure of five lyngbyabellin derivatives from a Papua New Guinea collection of the marine cyanobacterium Lyngbya majuscula. Tetrahedron 2005, 61, 11723–11729.
- Schmidt, E.W.; Nelson, J.T.; Rasko, D.A.; Sudek, S.; Eisen, J.A.; Haygood, M.G.; Ravel, J. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Natl. Acad. Sci. USA 2005, 102, 7315–7320, doi:10.1073/pnas.0501424102.
- Ziemert, N.; Ishida, K.; Quillardet, P.; Bouchier, C.; Hertweck, C.; de Marsac, N.T.; Dittmann, E. Microcyclamide biosynthesis in two strains of Microcystis aeruginosa: From structure to genes and vice versa. Environ. Microbiol. 2008, 74, 1791–1797, doi:10.1128/AEM.02392-07.
- Philmus, B.; Christiansen, G.; Yoshida, W.Y.; Hemscheidt, T.K. Post-translational modification in microviridin biosynthesis. Chembiochem 2008, 9, 3066–3073, doi:10.1002/cbic.200800560.
- Balskus, E.P.; Walsh, C.T. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 2010, 329, 1653–1656, doi:10.1126/science.1193637.
- Tan, L.T.; Chang, Y.Y.; Ashootosh, T. Besarhanamides A and B from the marine cyanobacterium Lyngbya majuscula. Phytochemistry 2008, 69, 2067–2069, doi:10.1016/j.phytochem.2008.04.021.
- Essack, M.; Alzubaidy, H.S.; Bajic, V.B.; Archer, J.A. Chemical compounds toxic to invertebrates isolated from marine cyanobacteria of potential relevance to the agricultural industry. Toxins (Basel) 2014, 6, 3058–3076, doi:10.3390/toxins6113058.
- Stewart, I.; Schluter, P.J.; Shaw, G.R. Cyanobacterial lipopolysaccharides and human health—A review. Health 2006, 5, 7, doi:10.1186/1476-069X-5-7.
- Chirasuwan, N.; Chaiklahan, R.; Ruengjitchatchawalya, M.; Bunnag, B.; Tanticharoen, M.J.A.; Resources, N. Anti HSV-1 activity of Spirulina platensis Kasetsart J. (Nat. Sci.) 2007, 41, 311–318.
- de Jesus Raposo, M.F.; De Morais, A.M.B.; De Morais, R.M.S.C. Marine polysaccharides from algae with potential biomedical applications. Drugs 2015, 13, 2967–3028.
- Delattre, C.; Pierre, G.; Laroche, C.; Michaud, P. Production, extraction and characterization of microalgal and cyanobacterial exopolysaccharides. Adv. 2016, 34, 1159–1179, doi:10.1016/j.biotechadv.2016.08.001.
- Moore, R.E. Toxins, anticancer agents, and tumor promoters from marine prokaryotes. Pure Appl. Chem. 1982, 54, 1919–1934.
- Banker, R.; Carmeli, S. Tenuecyclamides A−D, cyclic hexapeptides from the cyanobacterium Nostoc spongiaeforme tenue. J. Nat. Prod. 1998, 61, 1248–1251.
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87.
- Gershenzon, J.; Dudareva, N. The function of terpene natural products in the natural world. Chem. Biol. 2007, 3, 408–414.
- Pattanaik, B.; Lindberg, P. Terpenoids and their biosynthesis in cyanobacteria. Life 2015, 5, 269–293.
- Belin, B.J.; Busset, N.; Giraud, E.; Molinaro, A.; Silipo, A.; Newman, D.K. Hopanoid lipids: From membranes to plant–bacteria interactions. Rev. Microbiol. 2018, 16, 304.
- Takaichi, S.; Mochimaru, M. Carotenoids and carotenogenesis in cyanobacteria: Unique ketocarotenoids and carotenoid glycosides. Mol. Life Sci. 2007, 64, 2607.
- Prasanna, R.; Sood, A.; Jaiswal, P.; Nayak, S.; Gupta, V.; Chaudhary, V.; Joshi, M.; Natarajan, C. Rediscovering cyanobacteria as valuable sources of bioactive compounds. Biochem. Microbiol. 2010, 46, 119–134.
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. Microbiol. 2018, 56, 280–285, doi:10.1007/s12275-018-8014-6.
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313, doi:10.1093/bioinformatics/btu033.
- Mejean, A.; Mann, S.; Vassiliadis, G.; Lombard, B.; Loew, D.; Ploux, O. In vitro reconstitution of the first steps of anatoxin-a biosynthesis in Oscillatoria PCC 6506: From free L-proline to acyl carrier protein bound dehydroproline. Biochemistry 2010, 49, 103–113.
- Dittmann, E.; Gugger, M.; Sivonen, K.; Fewer, D.P. Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol. 2015, 23, 642–652.
- Balskus, E.P.; Walsh, C.T. Investigating the initial steps in the biosynthesis of cyanobacterial sunscreen scytonemin. Am. Chem. Soc. 2008, 130, 15260–15261.
- Ansari, M.Z.; Yadav, G.; Gokhale, R.S.; Mohanty, D. NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004, 32, W405–W413.
- Méjean, A.; Ploux, O. A genomic view of secondary metabolite production in cyanobacteria. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2013; Volume 65, pp. 189–234.
- Arnison, P.G.; Bibb, M.J.; Bierbaum, G.; Bowers, A.A.; Bugni, T.S.; Bulaj, G.; Camarero, J.A.; Campopiano, D.J.; Challis, G.L.; Clardy, J. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Prod. Rep. 2013, 30, 108–160.
- Montalbán-López, M.; Scott, T.A.; Ramesh, S.; Rahman, I.R.; van Heel, A.J.; Viel, J.H.; Bandarian, V.; Dittmann, E.; Genilloud, O.; Goto, Y. New developments in RiPP discovery, enzymology and engineering. Prod. Rep. 2020.
- Han, B.; McPhail, K.L.; Ligresti, A.; Di Marzo, V.; Gerwick, W.H. Semiplenamides A–G, Fatty acid amides from a Papua New Guinea collection of the marina cyanobacterium Lyngbya semiplena. Nat. Prod. 2003, 66, 1364–1368.
- Chi, Z.; Su, C.; Lu, W. A new exopolysaccharide produced by marine Cyanothece 113. Bioresour. Technol. 2007, 98, 1329–1332.
- Markou, G.; Nerantzis, E. Microalgae for high-value compounds and biofuels production: A review with focus on cultivation under stress conditions. Adv. 2013, 31, 1532–1542.
- Delattre, C.; Vijayalakshmi, M. Monolith enzymatic microreactor at the frontier of glycomic toward a new route for the production of bioactive oligosaccharides. Mol. Catal. B Enzym. 2009, 60, 97–105.
- Kraan, S. Algal polysaccharides, novel applications and outlook. In Carbohydrates-Comprehensive Studies on Glycobiology and Glycotechnology; IntechOpen: London, UK, 2012.
- Mišurcová, L.; Orsavová, J.; Vávra Ambrožová, J. Algal polysaccharides and health. Bioactivity Biotechnol. 2015, 1. 109–144.
- Skjånes, K.; Rebours, C.; Lindblad, P. Potential for green microalgae to produce hydrogen, pharmaceuticals and other high value products in a combined process. Rev. Biotechnol. 2013, 33, 172–215.
- Swain, S.S.; Paidesetty, S.K.; Padhy, R.N. Antibacterial, antifungal and antimycobacterial compounds from cyanobacteria. Pharmacother. 2017, 90, 760–776.