Definition (Draft for you)
CRISPR-Cas9 is a simple two-component system that allows researchers to accurately edit any sequence in the genome of an organism. This is achieved by the guide RNA, which recognizes the target sequence, and the CRISPR-associated endonuclease (Cas) that cuts the targeted sequence.
- CRISPR-Cas9
1. Introduction
CRISPR-Cas9 provides sequence-specific recognition tool as well as a simple technique to produce sequence-specific genomic breaks. In the past decade, CRISPR-Cas systems have transitioned from an adaptive immune defense system in bacteria and archaea to revolutionary genome-editing tools used in various organisms, from bacteria to plants and animals including humans [8]. The resulting CRISPR technologies have driven innovations for treating genetic modification and device to another dimension by modifying diverse eukaryotic genomes [9]. Also, several microalgae species have been modified with CRISPR-Cas genome editing such as Nannochloropsis oceanica [10], Phaeodactylum tricornutum [11], Thalassiosira pseudonana [12], and Chlorella vulgaris [13]. The current review solely focuses on the green algae C. reinhardtii.
2. Summary of the Pipeline for Using the CRISPR-Cas System for Genome Editing in Green Microalgae C. reinhardtii
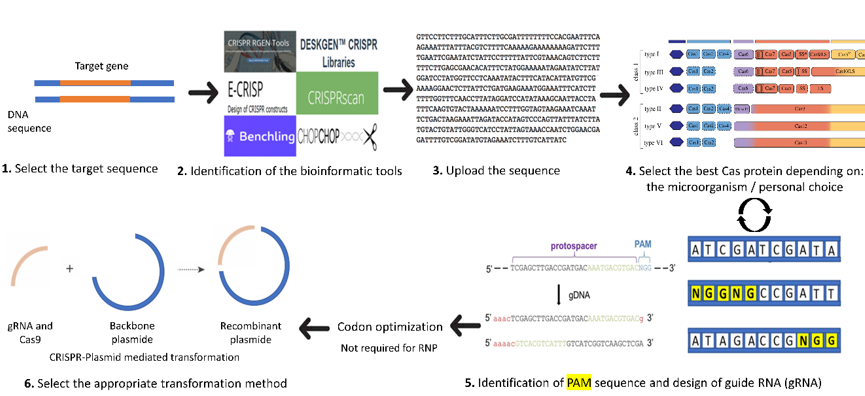
A brief introduction on the order of events to use in the design of a CRISPR knock-out or knock-down system in C. reinhardtii is presented in Figure 1. Details for each step are described in the following sections. The first step starts with the selection of the target gene, preferably a stable gene without multiple copies in the desired genome. Next comes the choice of the software to use and adapted to C. reinhardtii. The third step includes the design of the guide RNA by uploading the target gene sequence into the software and it also involves selecting the adequate Cas protein for the work (selected from the list usually generated by the software and depending on personal choice). The software will help to choose the appropriate PAM (protospacer adjacent motif) and will generate different single guide RNA (sgRNA) with highlighted PAM (Figure 1). The PAM is a short DNA sequence (usually 2–6 nucleotides in length) that follows the DNA region targeted for cleavage by the CRISPR system. The PAM is required for the Cas nuclease to cut and is generally found 3–4 nucleotides downstream from the cut site. Then, we can select the most suitable sgRNA for the desired project (with the minimum of off-target loci if possible).
Once the appropriate sgRNA, Cas protein, and targeted gene are selected, comes the selection of the transformation method. Select the proper CRISPR system for the transformation of either plasmid-mediated CRISPR or RiboNucleic Protein-CRISPR (RNP-CRISPR). If the RNP-CRISPR is selected, the optimization of Cas9 and synthesis of guide RNA can proceed with or without codon optimization. However, if the plasmid-mediated transformation is selected, the user should choose a backbone plasmid in which he has to clone or synthesize the sequence of Cas and gRNA. Then, optimize the codons and synthesize the sequences of Cas protein and sgRNA.
Figure 1. Important steps for the design of sgRNA (adapted from [24]). 1: Select the target gene sequence. 2: Identify the bioinformatic tool suitable for the organism (here, C. reinhardtii). 3. Upload the sequence of the gene of interest and select the organism (here in C. reinhardtii) in the database of the software. 4: Select the best Cas protein to be used or in some software the PAM sequence. 5: The software will automatically choose the adequate Cas protein identified by the user and its locations in the genome will be indicated by different colors or arrows. Once the PAM (highlighted in yellow) is selected, the user can choose the option design guide RNA and save the sequence. Then, Cas protein and gRNA can be codon optimized for better protein expression. 6: Select the proper CRISPR system for the transformation (either plasmid-mediated CRISPR or RNP-mediated CRISPR).
2.1. Codon Optimization
The lack of compatible parts between microorganisms is one of the most important reasons for the expansion of synthetic biology tools, especially codon optimization. Even though foreign genes are easily integrated into C. reinhardtii, the transgene expression is often weak [25]. The efficiency of the gene expression strongly depends on the nucleotide identity (rich or poor on GC content, especially at the wobble position (3rd letter of codon)) and depends on the usage of the codon by the microalgae [26]. The codon sequences (CDSs) of C. reinhardtii have an average of 68% GC content [27]. By consequence, a slight change in the GC content in C. reinhardtii codons drastically affects the gene expression at the chromatin level.
It is worth mentioning that previous data of codon usage comes from the web site Kazusa [28] and is calculated from 846 CDSs. However, nowadays, different software and companies are doing rapid codons optimization for C. reinhardtii sequences on the basis of the codon adaptation index (CAI). The CAI of each gene is computed as the geometric mean of the relative adaptiveness of all the codons in the coding sequence [26]. It is also worth highlighting that the addition of intron-containing sequence can enhance the expression in Chlamydomonas [29]. It might be an avenue in the improvement of its toolbox.
Beside the genetic cassettes (sgRNA) to be inserted, some of the Cas9 protein sequences used in previous cited articles were also codon optimized. Thus, different C. reinhardtii cell type has been modified genetically in regard of this tool. However, some issues regarding the efficiency of transformation have been encountered. The construction of a proficient and mature CRISPR-Cas expression system is now essential for efficient targeting.
This entry is adapted from the peer-reviewed paper 10.3390/life10110295