玉米因其营养特性和在食品工业中的广泛应用而在食品工业中很重要。同时,玉米是世界上产量(吨)最大的粮食作物[1],因此,它被许多国家视为国家粮食安全的战略作物[2].因此,对玉米产品的持续需求推动了其产量的增加。转基因技术可用于改变玉米的遗传性状,常用的性状转化方法很多[3][4][5][6].目前,通常研究的性状包括害虫和除草剂抗性。
1996年,美国首次批准Cry1Ab转基因玉米(“Bt176”、“MON810”和“Bt11”)的商业化,转基因玉米现已推广应用27年。[7].截至本出版物,已有244个转基因玉米品种被批准种植[8].根据国际农业生物技术应用服务社(ISAAA)的一份报告,截至29年,转基因作物在2019个国家种植。转基因玉米种植面积为6.09×107嗯22019年,占转基因作物种植总面积的32%。种植面积是美国最大的,为3.317×107嗯2,其次是巴西和阿根廷[9].转基因玉米在全球主要用于动物饲料[10][11]或作为工业原料提取酒精。只有一小部分被人类直接消费。用于食品的转基因玉米主要用于提取玉米油[12],用于制作玉米糖浆、玉米粉或其他玉米成分[13]尤其是玉米淀粉,在食品工业中广泛用作增稠剂、胶凝剂、填料、保水剂[14].用于食品的转基因玉米也直接用作食品生产和加工的原料,例如在墨西哥生产玉米饼[15];南非的白玉米,它是大多数人的主食[16],以及玉米片、爆米花和玉米相关零食[17].毫无疑问,转基因玉米已融入全世界人类的生活,因此,有效规范其生产和加工非常重要。
欧盟(EU)对市场上的转基因产品采用了可追溯性管理系统和强制性标签系统。1830年2003月生效的欧盟法规2004/0/EC规定,含量超过9.<>%(质量分数,%m/m)的产品必须标有“遗传改良”或“从转基因作物加工”的字样。此外,应使用DNA拷贝数的比例作为表达方法。以质量百分比为准标准物质的测定结果,拷贝数百分比(DNA拷贝数比,%cpT/cpE)的测量结果换算成质量百分比[18].在中国[19]北美洲[20][21], 澳大利亚, 新西兰[22]印度[23]等国家有法律法规明确转基因产品的标签管理体系。这确保了企业经营者和消费者能够获得准确的信息,以便他们能够有效地行使他们的选择自由,并能够控制和验证标签声明。后一项要求使得有必要通过可靠的检测方法检测转基因生物(GMO)的存在。转基因作物的检测按目标大致可分为核酸、蛋白质、代谢物的检测。在生物细胞中,与蛋白质相比,DNA相对稳定,即使在作物加工后也不容易被破坏,因此微量或可检测量的DNA片段可能残留在产品中。因此,DNA检测通常是识别作物转基因成分的首选方法。在常见的多核苷酸检测方法中,诸如启动子(例如花椰菜花叶病毒(CaMV)35S启动子等靶标[24])、终止子(例如诺帕林合酶终止子(T-nos) [25][26]),或标记基因(CP4-EPSPS和pat [27][28]) 通常作为转换的代理标记调用。然而,上述靶基因的检测方法无法区分转移相同外源基因的不同转基因作物品系,特异性较低。因此,跨越插入的转基因和侧翼DNA连接的一对引物通常用于识别转基因特异性事件[29][30],即转基因作物的品系,通过检测外源基因与植物基因组的连接区域。目前,该方法广泛用于鉴别转基因玉米品系,如MON810、NK603[31], Bt11, TC1507, GA21[32],等等。
一般来说,核酸检测需要经过一个核酸提取纯化的过程,靶标检测才能获得结果。提取的核酸的纯度和质量决定了后续扩增的有效性,靶标的设定是检测转基因玉米样品特异性的基础。获取结果的方法也是基于直觉和便利性,根据不同检测场景的需求。例如,简单快速的现场测试方法对于规范转基因作物的进出口至关重要。在这种情况下,收集代表性样本的方式以及分析的时间和可靠性对于顺利实施法规和市场监督至关重要。
2. 核酸提取技术
基于DNA的测定用于检查加工产品的转基因状态,但必须确保DNA的质量。在研究不同种类材料的分子生物学检测方法时,DNA提取方法的选择直接影响其性能和效用。目前,植物基因组DNA的常用提取方法是十二烷基硫酸钠(SDS)[33], 十六烷基三甲基溴化铵 (CTAB)[34],尿素提取法,Chelex-100法[35]、碱裂解法、聚乙烯吡咯烷酮-40 (PVP-40)[36]、高盐低pH萃取法[37]和商用套件[38]等。这些DNA提取方法在原则上通常是一致的,包括细胞裂解以释放DNA和去除杂质以进行DNA纯化(图1)。然而,不同植物材料中蛋白质、多糖、酚类等物质的含量并不相同,这给DNA的提取、分离和纯化造成了很大的困难。对于植物来源的样品,应根据样品的特性选择合适的方法,并且可能需要对方法进行调整和优化。
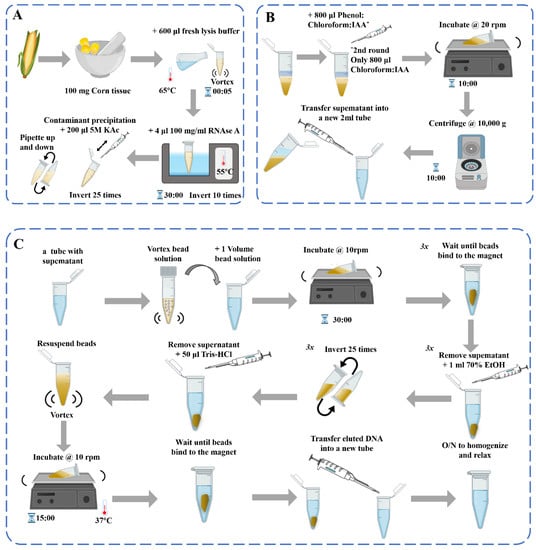
图1.DNA提取过程示意图[39]:(A)脱氧核糖核酸提取;(b)污染物沉淀和苯酚:氯仿纯化2×;(C)用磁珠纯化基因组DNA。
在玉米样品的DNA提取过程中,有针对各种植物组织的玉米原料的相关提取方法,如花粉、叶子、玉米丝[40]、胚乳等[41].玉米是一种被子植物,其果实具有独特的结构,主要由果皮、种皮、胚乳和胚胎(胚芽、胚根、下胚轴和子叶)组成。有性繁殖玉米的果皮和种皮是由雌亲的子房壁和外皮发育而来的,所有的遗传物质都来自雌性亲本。成熟玉米的果皮和种皮几乎一起生长,很难分离。胚乳由受精极核发育而来,其遗传物质与胚胎和成株不同;因此,在提取原料时应适当考虑。传统的基因筛选方法涉及从叶子中提取基因组DNA[42][43]在田间种植大量玉米后单个植物的(或嫩叶)。单籽玉米DNA提取方法主要涉及粉碎种子或利用种子胚乳提取DNA。玉米粗品的DNA提取方法主要用于饲料中转基因生物的检测[44][45].根据DNA降解程度由小到大,相关食物可分为生玉米、冷冻玉米、罐装玉米、干包玉米样品[46].深加工玉米产品主要是调味、膨化、油炸、糖化、发酵面粉产品,如玉米油[47]、玉米淀粉、玉米片、爆米花、玉米棒、脆玉米角、哇头[48]等。
细胞裂解,包括机械、酶和化学裂解,是从细胞组织中提取DNA的基本过程。机械开裂,如液氮研磨、热冲击、均质化、超声波处理等[49],可能导致DNA断裂,在实际应用中一般起到辅助作用。需要严格控制超声强度和间隔时间,以避免DNA过度断裂。酶裂解使用特定的酶(如果胶酶)来破坏细胞并释放核酸,这些核酸可以与化学裂解一起提取。
CTAB和SDS是两种最常用和最有效的化学裂解剂。通常,表面活性剂(如CTAB和SDS)倾向于与静电力,负轴力和疏水力驱动的聚合物(例如蛋白质和DNA)相互作用[50].CTAB能溶解细胞膜,在高盐(>0.7mol/L NaCl)溶液中,CTAB能与蛋白质和多糖形成可溶性稳定的复合物,但不能析出核酸[51].在低盐(0.1~0.5mol/L NaCl)溶液中[52],SDS可以在55~65°C的碱性条件下裂解细胞并使DNA游离,同时使蛋白质变性并与其结合[53].最近,离子液体(ILs)和磁性离子液体(MIL)已被探索为从复杂生物基质中提取DNA的新型溶剂[54].IL 和 MIL 通过阳离子和带负电荷的磷酸盐主链之间的静电相互作用以及溶剂和 DNA 碱基的烷基链之间的疏水相互作用促进 DNA 提取[55][56].微尺度电穿孔是一种释放细胞内材料的新兴技术[57].其作用机理是,当外加电脉冲的电场强度达到一定数量级时,细胞膜发生构型变化,出现大量微孔。这增加了细胞膜的通透性,有利于各种大分子物质(如DNA、RNA、蛋白质、化学小分子等)的释放。然而,调整施加的电压和脉冲长度以确定细胞裂解和DNA提取的最佳条件仍然需要实验验证。
粗 DNA 提取物含有大量蛋白质、RNA、糖和其他杂质;因此,DNA纯化是必不可少的。大多数蛋白质在用氯仿或苯酚处理后可以通过变性和沉淀去除,这是蛋白质去除的常用方法[58].苯酚和氯仿这两种不同的蛋白质变性剂交替使用,可以增强蛋白质去除的效果[49].应该注意的是,这些化学物质,苯酚和氯仿,具有一定的氧化性,如果使用不当会严重损害DNA。例如,鸟嘌呤对氧化特别敏感,接触苯酚/氯仿会导致形成8-氧鸟嘌呤[59].此外,苯酚和氯仿具有挥发性和毒性,根据美国卫生与公众服务部关于致癌物的报告,氯仿被归类为“根据实验动物研究中足够的致癌性证据,合理预期是人类致癌物”。[60].
RNA可在约1°C下用RNase A消化2~37 h除去,或者通过氯化铯密度梯度离心纯化DNA,从而得到高质量的DNA制备[58].玉米植物细胞具有增厚的次生壁和大液泡,储存大量次生物质,如多糖和多酚。表面活性剂CTAB在去除多糖方面优于SDS[61][62].适当增加CTAB的含量(根据实际情况,如提高到3%,但使用量不低于1%[63]β-巯基乙醇(0.2%–1%,根据实际情况确定)[64]能有效去除多糖等次生生物分子。同时,植物中所含的多酚在多酚氧化酶的催化作用下被氧化,导致提取的DNA质量降低。去除多酚作用的主要方法包括在提取介质中添加抗氧化剂或在研磨过程中添加PVP或抗坏血酸[65].通过使用较高浓度的盐和酸性较低的介质可以防止酚类化合物的进一步氧化[66].已经开发了带有二氧化硅基离心柱的商用固相萃取试剂盒,以标准化程序并使其更有效率。这些试剂盒使用含有CTAB或SDS的切割缓冲液,由解离盐组成的结合缓冲液,以促进DNA吸附到二氧化硅吸附剂上,以及含有有机溶剂的洗涤缓冲液来洗脱和纯化DNA[67].整个提取过程在室温下进行,实验要求严格性低,所得DNA的浓度和纯度满足大多数当代分子生物学应用的要求。目前,用于植物DNA提取的商业试剂盒也采用功能化磁性材料,通过使用外部磁铁简化纯化步骤。
对于转基因玉米的DNA提取,传统的CTAB方法是从高度加工的玉米面筋中提取可扩增DNA的最合适方法,这种面筋通常用作富含蛋白质的饲料成分。该方法可以在实验室测试中产生足够量的扩增DNA,以控制测试底物是否符合授权转基因玉米的事件容许限和标记阈值[44].与CTAB方法相比,SDS提取方法通常具有更高的DNA产量,更好的细胞裂解效率,更低的DNA剪切速率和更高的多样性[68].因此,在目前的众多DNA提取方法中,CTAB和SDS方法的新改进目前正在开发中。[53][69].
随着分子生物学、精准诊断和治疗领域的快速发展,对具有高通量、纯度和质量的新兴核酸提取技术的需求不断增加。磁珠(MBs)法作为一种简单、快速、可靠、自动化的核酸提取方法[70] for nucleic acid extraction has attracted more and more attention. The process of DNA extraction by the MBs method is simple, without repeated centrifugation, column separation, or vacuum filtration [71]. The entire process consists of four steps including lysis, binding, separation, and elution. Therefore, the process is fast and the extraction efficiency is high, culminating in extracted nucleic acids that are high in purity and concentration. Moreover, it is safe and non-toxic, does not use toxic reagents (such as phenol, etc.), reduces personnel hazards, and can be easily adapted for automated batch operation [72]. MBs and automatic nucleic acid extraction instruments can be used for high-throughput automated extraction of nucleic acids from large numbers of clinical samples. MBs with functionalized surfaces that can capture nucleic acids have been widely used to extract nucleic acids from biological samples, and multiple forms have been developed [73]. Examples include manual extraction using magnetic frames or microfluidic chips [74], automated robotic processing, and also the separation of ctDNA by superparamagnetic bead particles in microfluidic platforms for early cancer detection, etc. Combining them with traditional gene extraction methods can efficiently and quickly extract plant nucleic acids, and has an absolute advantage over other methods in the detection of low-content genetically modified components. Sebastian et al. [75] used centrifugal microfluidic technology, using continuous rotating magnetophoresis to facilitate magnetic bead integration and nucleic acid extraction where needed. This solution solves the drawbacks of magnetic bead-based solid-phase extraction that may cause nucleic acid loss due to the handling of magnetic beads when they are transferred from one chamber to another, resulting in higher yield and purity in the end. Jiang et al. [70] and others established a no-elution MB-based nucleic acid extraction method by introducing PEPPG F68 into the lysate and using NaOH solution instead of alcohol as the washing buffer. It avoids the dilution and loss of the target nucleic acid during the elution process, as well as the possible loss of sensitivity and false-negative results. At the same time, the detection sensitivity of loop-mediated isothermal amplification (LAMP) is significantly improved, which has broad application prospects. The current demand by molecular biologists is for convenient, rapid, and inexpensive DNA extraction and detection methods and more compact, portable equipment options to enhance real-time capabilities. The merging of extraction and microflow body technologies [76] to automate nucleic acid detection [77][78] is also in high demand.
3. Traditional Detection Technology
3.1. Variable-Temperature Amplification
Event-specific polymerase chain reaction (PCR) targeting unique sequences spanning the insert DNA and flanking genomic DNA has the highest level of specificity and is commonly used to confirm the identity and authorization status of GMO ingredients and to quantify GMO content [79]. DNA-based PCR methods are considered the most reliable and versatile techniques for the identification and quantification of GMOs, with the chief method being variable-temperature amplification. Variable-temperature amplification technology includes three steps, high-temperature denaturation, low-temperature annealing, and a suitable-temperature extension, and generally refers to PCR technology and its derivatives, such as real-time fluorescent PCR [80][81], droplet digital PCR (ddPCR) [82], nested or semi-nested PCR [83], multiplex PCR [84], etc.
The PCR method is the most commonly used molecular detection technique and is the standard method for detecting GMOs [85]. Standard qualitative PCR is the most commonly used PCR detection method. The mechanism is as follows: after the primer and template DNA are specifically combined in accordance with the principle of complementary base pairing, under the catalysis of Taq DNA polymerase, using deoxyribonucleotide triphosphates (dNTPs) as the raw material, a new DNA strand is synthesized according to the principle of semi-conservative replication. After “n” times of amplification, the total number of progeny DNA is 2n, and finally achieves a million-fold amplification of the number of target DNA fragments, which facilitates the detection of subsequent target DNA fragments [86] and, finally, achieves a million-fold amplification of the number of target DNA fragments, which facilitates the detection of subsequent target DNA fragments. Standard PCR is used for the amplification of transgenic crop genes owing to its simple operation, high efficiency, and low cost. Quantitative PCR (qPCR), based on a standard curve, is considered the gold standard technique for the analysis of GMOs because of its high sensitivity and good stability [87]. Real-time fluorescent qPCR [88] is a technology that adds fluorescent groups to the PCR reaction system and uses the accumulation of fluorescent signals to monitor the entire PCR reaction process in real time. This technology uses the strength of the fluorescent signal to determine the number of specific amplification products over time, and an unknown template is quantitatively analyzed using a standard curve. This technology can quantitatively analyze DNA templates and has the characteristics of high sensitivity, specificity, and reliability; low pollution; and timely and accurate detection. It can perform both absolute and relative quantifications and is widely used to inspect GM maize [89]. Various forms of qPCR are constantly being developed to meet the needs of practical applications, focusing on duplex and multiplex reactions to improve detection throughput and efficiency.
The concept of digital PCR (dPCR) was first proposed by Vogelstein et al. in 1999 [90]. In dPCR, the reaction mixture is divided into many individual reactions called partitions, and each reaction does not contain one or more copies of the target. Reads are partitioned as negative or positive at the endpoints, and DNA concentrations are calculated using a Poisson distribution [91]. The partitioning of reaction volumes using wells on a chip in microfluidics/chip-based dPCR [92] and droplets in emulsion/ddPCR [93] are the two main approaches. In cdPCR, reactions are divided into hundreds or thousands of chambers in a single plate or array. Many studies have used chip-based platforms, such as the microwell chip-based QuantStudio 12k flex dPCR and 3D dPCR (Life Technologies), for the detection of GMOs in the field [94][95]. The Constellation system (Formulatrix) is a plate-based microfluidic dPCR system that offers five-color multiplexing. The biggest difference between the cdPCR platforms is the number of partitions created per sample and the number of samples analyzed in one run [96]. ddPCR has a synergistic effect on droplet microfluidics. It improves the sensitivity of PCR at the single-molecule level by dividing tens of microliters of PCR mixture into tens of thousands of droplets and it can perform absolute quantification without a standard curve, thus avoiding the amplification efficiency bias observed in qPCR. It enables accurate target determination even at low copy numbers and can be significantly cost-effective when combined with multiplexing [97]. However, the droplet reaction generator used for ddPCR is bulky and complicated, which is an important limitation for its use in on-site detection. Thus far, microfluidic platforms for droplet generation using centrifugal forces, such as those utilizing ferrofluids, electromagnets [97], and surface acoustic waves [98]. Using the working principle of the indirect pressurization method, Park et al. [99] developed a pushbutton-activated microfluidic dropenser (droplet dispenser). Its use for sample preparation in ddPCR eliminates the need for benchtop droplet generators and automated pipetting, making ddPCR an on-site molecular diagnostic tool. In conclusion, dPCR has proven to be an effective tool for the quantification of maize and soybean GMOs and GMOs in complex matrix samples with precision and accuracy the same as or better than qPCR methods [100]. Recently developed multiplex dPCR methods [87][101] may be useful for analyzing samples containing multiple genetic modification events.
More types of PCR technologies are constantly being developed to meet the requirements for on-site rapid detection and high throughput. The use of an ultrafast PCR system can significantly reduce PCR run times and the number of reagents required for analysis. Therefore, ultrafast PCR systems have recently been studied and applied in various fields. The latest example of an ultrafast PCR system is a system used in rice detection [102]. The analysis principle is the same as that of real-time fluorescent qPCR, based on SYBR green [103], except that Evagreen dye is used as the intercalating dye instead of SYBR green. It requires 18% of the detection time of traditional PCR and 23% of the detection time of real-time PCR, and it can support small portable analyzers, thus providing a new strategy for the on-site detection of GM maize.
3.2. Isothermal Amplification
Isothermal nucleic acid amplification technology is used for nucleic acid amplification at a constant temperature. According to the different methods of single-stranded template formation, isothermal amplification can be divided into the following four categories: (1) strand-displacing DNA polymerase-mediated reactions such as Loop-Mediated Isothermal Amplification (LAMP) [104], Rolling Circle Amplification (RCA) [105], Cross-Primed Amplification (CPA) [106], Nucleic Acid Sequence-Based Amplification (NASBA [107]), and Multiple Displacement Amplification (MDA) [108]; (2) Enzymatic unwinding primer annealing reaction, such as Helicase-Dependent Amplification (HDA) [109], Recombinase Polymerase Amplification (RPA) [110], and Ligase Chain Reaction (LCR) [111]; (3) RNA transcription-based amplification, such as Transcription Mediated Amplification (TMA); and (4) Requires reactions assisted by single-strand cleavage enzymes, such as Strand Displacement Amplification (SDA) [112], and Isothermal Strand Displacement Amplification (iSDA) [113]. LAMP, RPA, and CPA are widely used for the detection of GM crops.
LAMP employs a DNA polymerase and a set of four specially designed primers that recognize a total of six different sequences in the target DNA. Internal primers containing the sequences of the sense and antisense strands of the target DNA initiate LAMP [104]. The product is a mixture of stem-loop DNA with stems of various sizes and cauliflower-like structures. Multiple loops are induced by annealing between alternating inverted repeats of the target sequence in the same strand. This enables simpler and more selective detection. For example, through a mechanism similar to multivalent antigen antibody interactions, the target sequence has a higher degree of specificity [104]. This method is insensitive to inhibitors, and can be used with crude DNA samples. LAMP is a simple and reliable GM detection method that can be performed on a thermal cycler, heating block, or portable constant temperature real-time amplification system. After the reaction is completed, using nucleic acid staining or fluorescent dyes, such as SYBR® Green and hydroxynaphthol blue, the LAMP products can be visualized and monitored using turbidity analysis or real-time LAMP [114]. Since the first report on the LAMP method in 2000, the number of studies using LAMP to detect GM ingredients has increased annually [115][116][117].
RPA technology was first proposed in 2006 by Piepenburg et al. [118]. In RPA, isothermal amplification of specific DNA fragments is achieved by the binding of reverse oligonucleotide primers to the template DNA and extension via DNA polymerase. It does not require the orientation of the primers to their complementary target sequences. RPA can be used to amplify cDNA generated by the reverse transcription of double-stranded DNA, single-stranded DNA, methylated DNA, RNA, or miRNA, and multiple reverse transcriptases are used for RPA [110]. When using RPA directly in milk [119] or seed powder [120], only thermal lysis, nuclease-free water lysis, or EzWayTM Direct PCR buffer are required to release the desired nucleic acid. With the assistance of a variety of enzymes, the in vitro amplification of nucleic acid can be completed at a constant temperature of 31–37 °C for 20 minutes [110]. Traditional in vitro nucleic acid amplification techniques do not have a rapid response, high sensitivity, high specificity, or low equipment dependence. Compared with SDA, RCA, and LAMP, RPA does not require an initial denaturation step to generate single-stranded (ss)DNA from double-stranded (ds)DNA targets, highlighting its suitability for use in the field [121]. In 2014, a commercial RPA kit launched by the British company, Twist DX [122], made the detection more convenient. Simultaneously, a variety of probes can be combined to expand the application range of RPA technology. RPA appears to be particularly well suited for multiplexing, where different targets can be verified with different efficiencies; however, it currently requires laborious optimization steps. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPER-associated protein [123], lateral flow assays [124][125]和微流体[126]为转基因的现场即时检测提供了额外的选择(图2)。
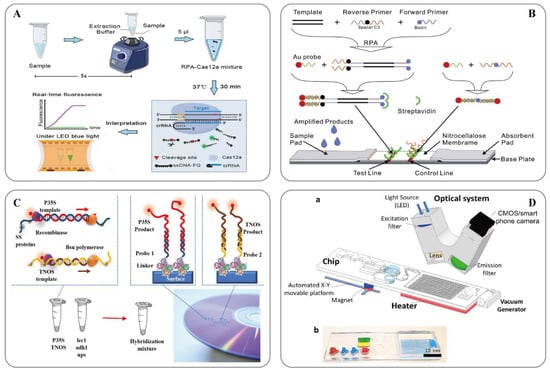
图2.(A) 用于快速和视觉核酸检测的ORCD检测系统示意图[123]; (B) LFNAA(侧流核酸测定)示意图[125]; (C) 基于多重 RPA 扩增的转基因生物检测方法方案(左)和阵列格式的杂交测定(右)[127]; (D) 用于样本到答案数字 RT-RPA 的微流控平台[126]: (a) 由磁性平台、加热器单元、真空发生器、带摄像头的检测部件和发光二极管 (LED) 组成的整个系统的示意图。一次性芯片有三个操作区域,用于制备和混合试剂以及检测病原体。(b) 用于芯片可视化的染料装载室。棕色、蓝色和红色分别表示用于样品制备的裂解室、洗涤室和洗脱室。绿色和黄色腔室分别显示RPA混合物和矿物油室。
CPA技术是由美塔生物科技有限公司开发的等温DNA扩增系统[106].系统仅依靠一个环结构进行复制[128].CPA测定能够在恒定温度下扩增核酸序列,并且只需要一种具有链置换活性的酶和一组五个引物即可进行CPA反应,而无需初始变性步骤或添加切口酶[129].在63 °C的测定温度下,DNA模板中瞬时自发变性气泡下引物模板杂交体的形成比相对于模板DNA的高浓度引物对模板链的重新退火更有利。通过退火与模板链不互补的5'末端的交叉引物以及交叉引物上游置换引物的结合来促进链置换。由此产生的靶DNA指数扩增具有高度特异性和灵敏度[106].传统上,CPA通过昂贵的荧光技术,繁琐的凝胶电泳程序或使用分光光度计测量浊度来产生结果。这些方法需要复杂而笨重的光学元件或暴露于致癌染料,这限制了它们在资源有限的实验室中的更广泛应用[128].此外,比色指示剂,如pH敏感染料(中性红)、孔雀石绿(MG)和羟基萘酚蓝(HNB)[130]也被用作监测CPA反应的补充技术。
3.3. 基因芯片技术
基因芯片技术是美国公司Affymetrix在1990年代开发的一种转基因食品检测技术。近年来,它作为一种高科技分子生物学工具迅速发展。基因芯片也称为DNA芯片或微阵列[131].该技术涉及在固相载体上以有序密度排列大量DNA片段或寡核苷酸片段,使用固相载体表面的特定探针与标记样品杂交,以及使用芯片扫描仪检测和分析杂交信号[132].它可以定性和定量地准确检测样品中不同类型的DNA序列。当微阵列杂交用于基因分析时,可以从很少的实验样品中获得有关基因表达差异的信息。当使用该技术进行检测时,对样品进行预处理和纯化以获得高纯度的DNA样品,然后通过PCR扩增,用荧光标记,并与基因芯片上的DNA探针杂交。然后读取信号以获得结果[133].基因芯片可以固定大量针对不同靶基因的寡核苷酸探针;因此,基因芯片技术可以同时检测数十甚至数百个基因[134]并且可以检测转基因食品中的多种成分。卢等.[135]采用基因芯片检测方法结合多重PCR,在单个芯片上同时检测多对基因。检测了七种转基因玉米成分:Bt176、Bt11、GA21、Mon810、Mon863、TC1507和NK603。该方法大大提高了检测的准确性和效率,灵敏度高达0.01%。图尔克等.[131]测试了1830种不同的探针,并为12个转基因品种(1个玉米和<>个大豆品种)开发了高密度寡核苷酸微阵列平台。该方法消除了PCR扩增步骤的需要,简化了分析并允许对每个检测到的GMO进行定量,从而能够以<>%(DNA浓度)的灵敏度对每种转基因作物进行特异性检测。
基因芯片技术具有高并行性、高通量、高特异性、高灵敏度和自动化等优势。但由于该技术开发起步较晚,综合性强,专业性强,成本高,制造基因芯片的过程相对复杂,存在一定的背景干扰等问题。因此,该技术在实际应用中的普及和推广受到限制[136].目前,全球种植的转基因食用农产品种类有限,转基因成分的检测往往只需要检测几十个靶基因,而基因芯片技术可以进行多基因甚至全基因检测。[134].因此,考虑到实验成本和利用率,该技术目前不适合转基因检测。一般来说,基因芯片技术需要极小的样本,具有快速、省时、无污染、准确、适合自动化操作等优点;然而,目前使用该技术的研究是在疾病诊断和微生物检测领域,转基因检测的研究更集中在微流控芯片上。[137].微流控芯片通过微处理技术将多个操作平台集成到一个芯片中,样品和试剂消耗少,反应速度快,并行工艺大量。因此,它具有更大的发展潜力。
