Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Oral cancer (OC) is among the most prevalent cancers in the world. Certain geographical areas are disproportionately affected by OC cases due to the regional differences in dietary habits, tobacco and alcohol consumption. However, conventional therapeutic methods do not yield satisfying treatment outcomes. Thus, there is an urgent need to understand the disease process and to develop diagnostic and therapeutic strategies for OC.
- head and neck cancer
- non-coding RNA
- cancer diagnosis
- cancer therapy
- microRNA (miRNA)
- long non-coding RNA (lncRNA)
- Circular RNAs (circRNA)
1. Introduction
Oral squamous cell carcinoma (OSCC) or oral cancer (OC) is the most prevalent type of head and neck cancer that arises in the tongue, lips, and floor of the mouth. In the year 2020, GLOBOCAN estimated around 377,713 total cases and 177,757 deaths worldwide from lip and oral cavity cancer, in which India alone showed a high burden of the disease [1]. The American Cancer Society has recently estimated the incidence of cancers in the oral cavity and pharynx, with around 54,000 new cases and about 11,580 deaths in the year 2023 alone [2]. Some of the most common factors with which OC progression is often linked include high tobacco and alcohol consumption, as well as an infection caused by human papillomavirus (HPV) [3]. Despite the advancement of conventional therapeutic strategies, the overall survival rate of OC is barely 50%, and even worse in the case of metastasis [4]. Since 1991, the USA has shown a continuous decline in overall mortality by 33%. However, the mortality rate of OC has shown a continued increase by 2% in men and 1% in women per year [2]. The aggressiveness and heterogeneity of the disease with delayed diagnosis, lack of early detection markers, lack of effective chemotherapeutic drugs, therapy resistance, and side effects often make the management of the disease complicated [5]. Although the U.S. Food and Drug Administration (FDA) approved EGFR targeted therapy and PD-1/PD-L1 immune therapy show some promising results, but these have achieved limited success [6][7]. Thus, there is an urgent need to understand the disease process and to develop better diagnostic and therapeutic strategies.
Recent developments in high throughput genome sequencing have revealed that more than 90% of the human genome encodes non-coding transcripts that do not code for any protein [8]. For a long while, non-coding RNAs (ncRNAs) were considered junk materials for cells. Recent knowledge on comprehensive molecular evaluations has led to the noncoding genes acquiring considerable attention nowadays in the advancement of several diseases, including cancers. Based on structural and functional characteristics, types of ncRNAs present in a cell are tRNA, rRNA, long non-coding RNA (lncRNA), circular RNA (circRNA), microRNA (miRNA), PIWI-interacting RNA (piRNA), and small nucleolar RNA (snoRNA) (Figure 1). The ncRNAs are sometimes tissue-specific and cellular-compartment-specific in their distribution and expression, and sometimes ubiquitous. NcRNAs are found to interact with nucleic acids or proteins to alter their conformation, activities, and stabilities. The differential expressions of ncRNAs are reported in different cancers. For example, miR-15/16, miR-29, miR-34, miR-200 family, let-7, miR-21, miR-155, miR-17–92 cluster, miR-221/222, miR-195 and miR-26b were significantly modulated in different cancers, including leukemia, prostate cancer, colorectal cancer, pancreatic cancer, liver cancer, lung cancer, breast cancer, glioblastoma, ovarian cancer, renal cancer, and thyroid cancer, and involved in cancer progression, metastasis, and drug resistance [9][10]. The high expression of miR-195 and miR-26b and down-regulation of their common target gene Semaphorin 6D (SEMA6D) were found to be associated with therapy resistance in breast cancer and, thus, this signaling axis is suggested as a predictive marker for chemotherapy response [10]. Likewise, up-regulation of lncRNA HOTAIR and MALAT1, down-regulation of lncRNA Meg3 and dual function of lnRNA H19 were seen in different cancer types, including lung cancer, ovarian cancer, prostate cancer, breast cancer, colorectal cancer, gastric cancer, and liver cancer [9]. Among the circRNAs, circPRKCI and circHIPK3 were found to be up-regulated in glioma, lung cancer, breast cancer, colorectal cancer, gallbladder cancer, gastric cancer, and ovarian cancer [9]. Differential expressions of piR-651, piR-823, piR-932 were reported in cancers of lung, breast, colorectal, esophageal, and gastric cancer [9]. Apart from the primary tissues, differential expressions of the ncRNAs in body fluid, including blood and saliva, suggest their importance as diagnostic and therapeutic biomarkers.
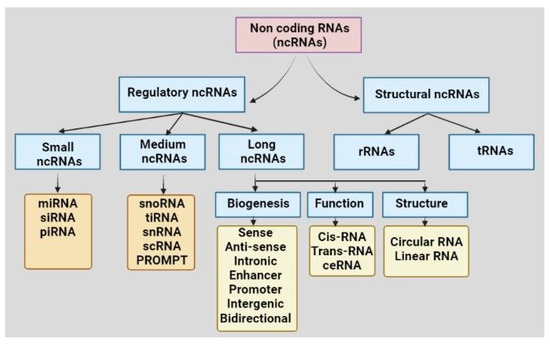
Figure 1. Classification of non-coding RNAs based on structures and functions. Abbreviations: non-coding RNA: ncRNA, rRNA: ribosomal RNA, tRNA: transfer RNA, miRNA: microRNA, piRNA: PIWI-interacting RNA, siRNA: small interfering RNA, snoRNA: small nucleolar RNA, tiRNA: transcription initiation RNA, snRNA: small nuclear RNA, scRNA: small cytoplasmic RNA, ceRNA: competing endogenous RNA, PROMPT: promoter upstream transcript.
To date, RNAseq studies have revealed more than 200 ncRNAs (including miRNA, lncRNA, circRNA, snoRNA, and piRNA) to have an association with OC progression. Researchers have described the types of ncRNAs, their functions, and their possible role as diagnostic and prognostic markers in OC.
2. Oral Cancer: Current Diagnostic and Prognostic Markers
Tissue biopsy and histological evaluation are the gold standard for diagnosing oral cancer (OC). However, this technique is painful for patients and causes a delayed diagnosis [11]. As non-invasive methods, metachromasia using iodine staining, or toluidine blue, which stains cancerous lesions, chemiluminescence-based lumenoscopy, auto-fluorescence based techniques like laser-induced auto-fluorescence (LIAF) or visually enhanced lesion scope (Velscope), and optical coherence tomography (OCT) provide assistance in early diagnosis and identification of oral pathophysiologic lesions [11]. The advancement of complete human genome insight, and the numerous potentials of cellular and epigenetic research, can be utilized as prognostic and diagnostic techniques for conducting rapid evaluation and treatment of oral lesions. The molecular diagnostic measures are divided into two types: nucleic acid-associated markers and protein-associated markers. In recent years, several biomarkers have been identified that could be utilized for the prognosis, diagnosis, differential diagnosis, prediction of recurrence, distant metastasis, and chemotherapy or radiotherapy resistance of OC. The biomarkers in OC include up-regulated expression of epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR), matrix metalloproteinases (MMPs), proliferating cell nuclear antigen (PCNA), Ki-67, Cyclin D1, Cathepsin-d, CD44, cytokeratins, p53 and p16 mutation or expression status, and the prevalence of human papilloma virus (HPV) and its oncogenes [12]. These biomarkers can aid in screening and early detection and prognosis of OC, which can improve treatment outcomes by allowing early intervention. However, these biomarkers are not enough as a primary diagnostic tool, and can only be useful in combination with other diagnostic methods to confirm malignancy and its stage. In addition, these markers are often not organ-specific. Therapeutic intervention by targeting these biomarkers shows limited efficacy and side effects.
3. Diverse ncRNAs in OC
3.1. Role of miRNAs in Oral Cancer
The effect of microRNAs (miRNAs) has gained considerable attention in the past few years for regulating the biological processes of multiple malignancies. The miRNAs are 19–25 nucleotide small RNA molecules that mainly interact with the 3′UTR of the target mRNA, and thus regulate gene expression. However, interactions with 5′UTR, promoter, and coding region have also been reported [13]. The miRNA can function as an oncogene or tumor suppressor gene, and the deregulation of miRNAs is seen in many cancers, including OC. Defects in miRNA biogenesis machinery, alterations in miRNA genes or transcriptional regulation, or epigenetic regulations are often associated with the miRNA deregulation mechanism.
With the aim of improving patient survival, numerous miRNAs have been discovered to have roles in the etiology of OC throughout time. The miRNAs associated with tissue, or bio-fluids like blood, and saliva, are found to regulate cancer growth, survival, invasion, metastasis, angiogenesis, chemotherapy, and radiotherapy resistance [14]. Simultaneous suppression of oncomiRs and replacement of tumor suppressor miRNAs are suggested to be an effective approach in designing an OC treatment strategy. OncomiRs like miR-21, miR-155, miR-196, miR-1237, miR-31, miR-455, miR-181, miR-184, miR-134, miR-146, miR-93, miR-372, miR-373, miR-103a-3p, miR-454, miR-654-5p, miR-188-5p, miR-626, miR-4513, miR-944, and miR-650 are found to be up-regulated in OC cell lines and patient samples. Many of the oncomiRs are found to be associated with OC diagnosis or prognosis, and some are suggested as biomarkers. The functions of the oncomiRs are associated with tumor progression, cell migration, drug resistance, inhibition of apoptosis, and induction of metastasis in OC. On the other hand, miRNAs such as miR-204, miR125b, miR-9, miR-26a/b, miR-491-5p, miR-375, miR-320, miR-218, miR-205, miR-181a, miR-138, miR-124, miR-99a, miR-34, miR-29a, miR-17, etc., function like tumor suppressors and are found to be down-regulated in OC cell lines, and patient samples. The miRNAs generally regulate the expression of oncogenes and tumor suppressors to modulate OC progression. For example, the miR-21 is one of the well-studied oncomiRs, and targets many tumor suppressor genes. In OC, the miR-21 interacts and regulates PTEN, RECK, PDCD4, TPM1, DKK2, and CADM1, resulting in induction of proliferation, invasion, and chemoresistance [15][16][17]. The transcription factor AP-1 (activating protein-1) activates miR-21 in various cancers [18]. Exposure of tobacco-smoke-associated nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) induces miR-21 and miR-155 expressions in lung and OC cell lines [19]. The miR-155 is involved in development of many cancers, and can act as both oncogene and tumor suppressor gene. In OC, the miR-155 is up-regulated as oncomiR, and associated with OC progression, metastasis, and drug resistance [20][21][22]. The transforming growth factor β (TGF-β)/Smad4 signaling activates miR-155 expression and promoter activity [23]. The TGF-β, a pleiotropic cytokine, regulates tumor suppression or promoting activity in OC [24]. Overexpression of TGF-β induces metastasis and immune modulation in OC [24]. Another well-studied miRNA is Let-7, which is responsible for cell differentiation in normal cells, and is found to become suppressed in OC [25]. The RNA binding protein Lin28A and B inhibit Let-7 biogenesis by binding at pre-miRNA gene [26]. In addition, lncRNA ROR, lncRNA H19, lncRNA PVT-1, and circ_CPA4, circ_HMCU act as miRNA sponges, and negatively regulate Let-7 expression in cancers [26]. Down-regulation of this miRNA increases expression of Twist and Snail which, in turn, promotes epithelial-to-mesenchymal transition (EMT) and plays a significant role in chemoresistance-to-cisplatin transition, as well as 5-fluorouracil (5-FU) [27]. The miR-124 is found to be deregulated in many cancers, including OC, due to promoter methylation. Overexpression of miR-124 inhibited integrin beta-1 (ITGβ1) and, thus, reduced OC migration [28]. Thus, the difference in the expression profiles gives both oncoMiRs and tumor suppressive miRNAs a role of potent biomarkers or therapeutic targets that might be beneficial to diagnose and treat OC.
3.2. Role of Long Non-Coding RNAs in Oral Cancer
Long non coding RNAs (LncRNAs) are more than 200 nucleotides long and they have similar biogenesis to messenger RNAs (mRNAs). They are transcribed by RNA-Polymerase II (Pol II), and undergo polyadenylation, splicing, and 5′-capping [29]. LncRNAs are localized either in the nucleus, cytoplasm, or both. Depending on the localization, they perform a variety of cellular processes by interacting with DNAs, RNAs, and proteins such as epigenetic modification, transcriptional regulation, RNA splicing, mRNA stabilization, translational regulation, sequestration of proteins, protein stabilization, and miRNA sponges [29][30][31][32]. Due to these regulatory functions, lncRNAs are associated with numerous disorders such as diabetes, inflammatory bowel diseases, AIDS, neurodegenerative diseases, different blood-related disorders, as well as cancers [33][34].
The lncRNAs exhibit aberrant expression patterns in OC, and play a significant role in the advancement of the disease [35]. In OC, the lncRNAs may either act as oncogenes or as tumor suppressor genes. The functions of some reported lncRNAs are summarized. Oncogenic lncRNAs, such as lncRNAs MALAT1, NEAT1, PVT1, ELDR, DLEU1, HOTAIR, HIFCAR, PCAT1, DANCR, etc., are associated with tumor proliferation, invasion, metastasis, angiogenesis, and drug resistance. For example, high MALAT1 expression is correlated with OC recurrence and metastasis [36]. It has also been demonstrated that MALAT1 promotes cellular proliferation and metastasis via controlling multiple signaling events, including Wnt/β-catenin signaling and PI3K/AKT/mTOR signaling pathways [36]. MALAT-1 is present in circulation, and shows importance as a serum biomarker. The MALAT1 is associated with resistance in multiple drugs, such as cisplatin, 5-fluorouracil, and paclitaxel [36]. Currently, the lncRNA MALAT1 is in OC clinical trial. The MALAT1 promoter contains transcription factors SP1 and SP3 binding sites [37]. Cooperative function of SP1 and SP3 activates MALAT1 expression [37]. In addition, STAT3/ TGF-β signaling axis, hypoxia-inducible factor (HIF)-2α, Yes-associated oncoprotein (YAP)-1, Jumonji C-domain–containing protein (JMJD)-1A, Octamer-binding transcription factor (OCT)-4, chemokine (C-C motif) ligand (CCL)-5, β-catenin, and Lysine-specific demethylase (KDM)-5B are involved in up-regulation of MALAT1 in OC and other cancers [37]. Another lncRNA ELDR is found to be up-regulated in OC cell lines and patient samples [38]. The ELDR inhibits miRNA-7, resulting in stabilization of EGFR. In addition, the ELDR interacts with RNA binding protein Interleukin Enhancer Binding Factor 3 (ILF3), and stabilizes cell cycle gene Cyclin E1. Interestingly, targeted inhibition of ELDR could inhibit in vivo tumor growth in a mouse model [38]. Overexpression of ELDR in normal oral keratinocytes (NOKs) induces cell proliferation and G2/M cell cycle progression through activation of CTCF/FOXM1/AURKA axis showing importance of the lncRNA as one of the OC driver genes [39]. It is not clear why the ELDR is up-regulated in OC; however, around 11% of samples contain the gene amplification in TCGA database (www.cbioportal.org, accessed on 28 June 2023).
On the other hand, the lncRNAs MEG3, GAS5, FENDRR, and PTCSC3 are down-regulated in OC. For example, the down-regulation of the lncRNA GAS5 increases miR-21 expression and helps in proliferation, invasion, EMT, and migration [40]. MEG3, being another tumor-suppressive lncRNA, suppresses miR-421 and the Wnt/β-catenin pathway [41][42]. Down-regulation of the lncRNA helps in the induction of the cell cycle, cell proliferation, metastasis, and suppression of cell apoptosis. Promoter methylation of MEG3 and GAS5 is one of the mechanisms of down-regulation in cancers [43][44].
3.3. Role of Circular RNAs (circRNA) in Oral Cancer
CircRNAs are closed-loop, extremely stable ncRNA molecules with no 3′ or 5′ ends and a poly (A) tail. They belong to the lncRNA kingdom, and have a higher half-life compared to linear RNAs [45][46]. Back-splicing and exon skipping of pre-mRNAs are the two processes by which circRNAs are produced. The transcription process is carried out via RNA pol II [47]. These RNA structures have the ability to withstand exonucleolytic breakdown by RNase R. It has been seen that 80% of circRNAs are present in the cytoplasm. However, the presence of circRNAs in the nucleus makes the control of gene expression possible [48].
Due to their closed-loop orientation, tissue specificity, high stability, and conservation, they serve as significant biomarkers for several diseases. These RNA molecules perform several regulatory functions, such as miRNA sponging, direct protein binding, and certain circRNAs are even translated into proteins [49][50]. Numerous studies have revealed that the majority of aberrantly expressed circRNAs play a significant role in controlling the progression of cancer by influencing a number of cancer hallmarks. Proliferative signaling, encouraging tumor and antitumor immunity, triggering angiogenesis, promoting invasion, metastasis, and deregulating cellular energetics are some functions that are associated with an aberrant circRNA profile [51].
Research on circRNA in OC has increased in recent years, and it has been discovered that these RNA molecules have a significant influence on the development, management, and prognosis of OC [52]. Like miRNA and lncRNA, these RNA molecules also play a role as both oncogenes and tumor suppressors in OC. For example, circ_0002185, circ_PVT1, circ_100290, circ_0001742, circ_HIPK3, circ_0001971, circ_DOCK1, circ_FLNA, circ_GOLPH3, circ_CLK3, circ_CDR1, circ_0014359, circ_LPAR3, circ_SEPT9, etc., are up-regulated, whereas circ_0000140, circ-PKD2, circ_0005379, circ_0004491, circ_SPATA6, circ_0086414, circ_0008309, circGDI2, circ_0007059, etc., are down-regulated in OC. The mechanism of differential expression of the circRNA in OC is not clear. The oncogenic circular RNA Circ_100290 acts as competing endogenous RNA (ceRNA), and inhibits miR-378a mediated suppression of glucose transporter GLUT1, resulting in the induction of glycolysis and cell growth [53]. The circ_PVT1 is derived from exon 3 of the oncogene of lncRNA PVT1 [54]. Recently, it was discovered that the mutant p53/ YAP/ TEAD transcription-competent complex is responsible for the up-regulation of circ_PVT1 in head and neck squamous cancer [54]. Through sponging miR-125b, Circ_PVT1 worked as a competitive endogenous RNA (ceRNA) to induce STAT3 signaling and cell proliferation [54]. Another circular RNA, called Circ_CDR1, has been stated to encourage autophagy under the hypoxic condition to enhance cell survival in OC, via control of the AKT/ERK-1/2/mTOR signaling pathway [55]. On the contrary, a study showed the association between suppressed expression of circ_0007059 and OC, via regulation of the AKT/mTOR pathway [56]. A high throughput sequencing study of OC samples identified significant down-regulation of circ_0005379 as compared to the adjacent normal tissues [57]. Up-regulation of circ_0005379 enhances cetuximab sensitivity, efficiently reduces OC proliferation, migration, invasion, and angiogenesis in vitro, and slows tumor growth in nude mice via inhibiting EGFR signaling [57]. All these studies suggest that these RNA molecules regulate OC via control over the major signaling pathways like MAPK, WNT/β-catenin, Notch, VEGF, and PI3K/AKT in OC [58]. The aggressive trait of OC was found to be linked to circ_PKD2 down-regulation. Overexpression of circ_PKD2 induces cell cycle arrest and apoptosis, and inhibited proliferation, migration, and invasion of OC through inhibiting miR-204-3p [59]. All these studies indicate the potential role of circRNA in OC.
3.4. Role of Small Nucleolar RNA (SnoRNA) in Oral Cancer
SnoRNAs are one of the many classes of non-coding RNA molecules present in the body. There are around 300 snoRNA sequences identified in the human genome. Although, snoRNAs are small in size, they are present in large quantities within the nucleus of cells [60]. Most snoRNAs are expressed in the intron of both coding and non-coding genes, while the remaining gets transcribed by RNA polymerase II (RNA pol II). Splicing, debranching, and co-transcription are the stages involved in their biogenesis. They play diverse roles in the development of ribosomal, small nuclear, and other pre-mRNA molecules via endonucleolytic disintegration and post-transcriptional regulation [60]. They also have the ability to control gene expressions by modifying and splicing mRNA. The snoRNAs form small nucleolar ribonucleoprotein complexes (snoRNP complexes) by binding to protein molecules, which then leads to the modification of rRNA bases [60].
SnoRNAs have roles in a variety of pathological and physiological processes. Studies have shown that snoRNAs control tumor growth, invasion, and metastasis, as well as cell death during the carcinogenesis process. More importantly, snoRNAs play a significant role in the development of OC tumors. Today, the differential expression of snoRNAs in OCs leads to the possibility of them being used as diagnostic and prognostic biomarkers [61]. However, the mechanism of differential expression of snoRNA in OC is not known clearly. Alteration in snoRNA biogenesis and post-transcriptional regulation may be involved in differential expression of different snoRNAs in OC. An in silico investigation using RNA seq data of 567 samples from the TCGA head and neck cancer cohort could identify 113 snoRNAs using p < 0 .05 as the cut-off [62]. The top significantly modulated snoRNAs were associated with DNA template regulation, RNA editing, regulation of cell proliferation, adhesion, invasion, metastasis, PI3K-AKT signaling, EMT, and angiogenesis pathways. Further analysis with the top five snoRNAs (SNORD114-17: ENSG00000201569, SNORA36B: ENSG00000222370, SNORD78: ENSG00000212378, U3: ENSG00000212182, and U3: ENSG00000212195) showed association with patient survival, indicating the importance of snoRNAs in disease progression and as biomarkers in OC [62]. A microarray analysis from eight OC samples identified 16 significantly modulated snoRNAs as compared to control samples; among them, 15 were significantly down-regulated and associated with patient survival [61]. The SNHG3, a snoRNA that gets up-regulated in OC patients, induces migration and cell proliferation of oral squamous cells. It targets the nuclear transcription factor-Y subunit gamma (NFYC) via the SNHG3/ miR-2682-5p axis and functions as a biomarker [63][64]. SnoRNA SNHG15 also gets overexpressed in OC cell lines, and facilitates the malignant behaviors of OC via miR-188-5p/ DAAM1 as a target [65]. Thus, snoRNAs play a role in assisting tumor growth in OC.
3.5. Role of piRNAs in Oral Cancer
PIWI-interacting RNAs (piRNAs) are a subclass of ncRNAs that can be divided into three primary categories: transposon-derived piRNAs, mRNA-derived piRNAs, and lncRNA-derived piRNAs. They are 24–31 nucleotides long, have 5′-end uridine or 10th position adenosine bias, and lack proper secondary structural features [66]. The piRNAs, composed of an array of different nucleotide sequences, are single-stranded ncRNAs that interact with P-element-induced wimpy testis (PIWI) proteins [67]. They are the largest group of ncRNAs, and are multi-functional. PiRNAs are instrumental in genome rearrangement, spermiogenesis, protein regulation, transposon silencing, epigenetic regulation, and germ stem-cell maintenance by binding to PIWI proteins to make a piRNA/PIWI complex [68].
The piRNAs are mostly known to be expressed in germ cells; however, their existence is also observed in cancer cells. Therefore, the question of employing these RNA molecules as a prognostic marker or therapeutic target arises. Current research has given evidence of the piRNAs/PIWI complex being used for the occurrence, development, metastasis, and recurrence of breast cancer [69] and lung cancer [70]. Some piRNAs have been found to have a role in the development of OC, and can be a potential biomarker or therapeutic target for OC in the future [71]. In the OC mouse model, piR354, piR415, piR832, and piR1584, have been found to interact with mRNA molecules [72]. Longer survival of patients with head and neck cancer is associated with low levels of piR-58510 and piR-35373 [73]. It is observed that genes like GALNT6, SPEDF, and MYBL2 that are paired with piRNAs are responsible for the suppression or progression of several OC tumors [72]. An in silico analysis using the RNA sequencing data of 455 head and neck cancer samples, and 43 matched non-tumors from The Cancer Genome Atlas (TCGA), showed a total of 305 piRNAs in both tumor and non-tumor tissues [74]. Among a total of 247 significantly altered genes, 25 piRNAs were exclusively expressed in non-tumor samples, and 87 were only expressed in tumors. The significantly up-regulated piRNAs, including the topmost gene FR140858, were associated with poor patient survival. This indicates the importance of piRNAs in OC as diagnostic and prognostic biomarkers [74]. Another study identified a panel of 30 piRNAs in 77 HPV positive head and neck cancer samples from the TCGA RNA seq data [75]. Simultaneous validation in cell lines further reported key piRNAs NONHSAT077364, NONHSAT102574, and NONHSAT128479 in HPV associated head and neck cancer development. Based on analysis of the tongue cancer GEO database (GSE196674 and GSE196688), 406 differentially expressed piRNAs were identified [76]. Further investigation identified a down-regulated piRNA: piR-33422 and its association with mevalonate/ cholesterol-pathway-related gene FDFT1 in tongue cancer. Using TCGA RNA seq data of 256 smoking-related head and neck cancer samples, a panel of 13 piRNAs were identified [77]. Among them, NONHSAT123636 and NONHSAT113708 were found to be associated with tumor stage, NONHSAT067200 with patient survival, and 6 other piRNAs with TP53 mutation and 3q26, 8q24, and 11q13 amplification. Further studies are needed to know the regulation of their expression and functional mechanism of piRNAs in OC.
4. NcRNAs in Oral Cancer Progression
OSCC or OC is a multistep process originating from epithelial cells by progressive accumulation of genetic and epigenetic alterations. The histological changes that occur during the carcinogenesis begin with atypical squamous cell hyperplasia to carcinoma in situ (CIS) through stages of dysplasia [78]. The molecular events associated with alterations in different protein coding genes during the development of OC are extensively studied [78]. However, the role of ncRNAs is not well-studied in this regard. Few studies have reported the potential modulation of miRNAs, lncRNAs, and circRNAs in premalignant lesions, including oral leukoplakia (LK), oral lichen planus (OLP), oral submucous fibrosis (OSF), and oral dysplasia, with respect to OC (Figure 2).
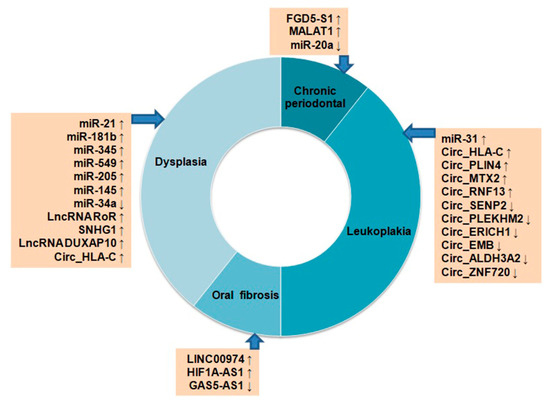
Figure 2. Differential expression of non-coding RNAs in oral pre-cancerous lesions. Up ↑ and down ↓ arrows indicate their up- or down-regulation status.
Up-regulation of miR-7, miR-31, miR-1293, and down-regulation of miR-133a, miR-204 and miR-206 were reported in OC samples. Among these miRNAs, significantly high expressions of miR-31 and down-regulation of its target gene C-X-C motif chemokine ligand 12 (CXCL12) were seen in LK and OLP tissues, suggesting their importance in OC progression from pre-cancerous stages [79]. The miR-21, miR-181b, miR-345, miR-549 and miR-205, were found to be overexpressed both in progressive dysplasia and OC [80]. Another study reported up-regulation of miR-145, lncRNA RoR, and SNHG1 and down-regulation of miR-34a from low-grade to high-grade dysplasia and, finally, to OC during carcinogenesis [81]. The lncRNA FGD5-AS1 inhibits NF-kB signaling, and gets down-regulated in chronic periodontal samples as compared to the healthy tissues [82]. On the other hand, lncRNA MALAT1 was found to be up-regulated in periodontal samples, and induced inflammation through TLR4 by targeting miR-20a [82]. In OSF, up-regulation of lncRNA LINC00974, HIF1A-AS1, and down-regulation of GAS5-AS1 were seen during the development of OSF [82]. An in silico study examined microarray data of 167 OC, 17 dysplasia, and 45 normal oral tissues from the GEO database for expression analysis of lncRNAs [83]. Among these groups, 200 lncRNAs were found to be common in three groups, and 1206 genes are common in OC vs. dysplasia groups. The differentially expressed genes (DEGs) identified among the three groups were found to regulate OC development through PI3K–Akt signaling and NF-kB signaling. Among the DEGs, lncRNA DUXAP10 is relatively new, and associated with the progression of OC development [83]. A high throughput sequencing study identified 366 significantly modulated circRNAs, including 65 up-regulated and 301 down-regulated in LK tissues as compared to the normal mucosa indicating their importance in OC development [84]. Some of the top significantly up-regulated circRNAs were Circ_HLA-C, Circ_PLIN4, Circ_MTX2, Circ_RNF13, and the down-regulated ones were Circ_SENP2, Circ_PLEKHM2, Circ_ERICH1, Circ_EMB, Circ_ALDH3A2, and Circ_ZNF720. The Circ_HLA-C showed stage-wise up-regulation from mild to severe dysplasia [84]. All these studies indicate the importance of ncRNAs in OC development from precancerous lesions to the most aggressive form; however, more mechanistic investigation is needed in this regard.
5. NcRNAs in Body Fluid and Exosomes of Oral Cancer as Diagnostic Markers
Even though there has been a lot of improvement in the treatment of OC in the last few years, the prognosis for OC remains poor. Involvement of extracellular vesicles or exosomes in biofluids, like blood and saliva, is seen in disease progression, cellular communication, and metastasis of many cancer types, including OC [85][86][87]. Moreover, studies have revealed that biomarkers in the blood are more intriguing, due to their low invasiveness and increased stability. In a recent study, aberrant expressions of 18 different circulating miRNAs have been discovered that possess a direct association with a poor prognosis for head and neck cancer [88]. Salivary exosomal miRNA-1307-5p was seen as a potent prognostic indicator for oral malignancies, as it has shown the ability to indicate poor prognosis as well as poor patient outcomes [89]. Increased levels of lncRNA TIRY derived from exosomes have been found to reduce miR-14 expression levels which, in turn, enhances OC progression and metastasis [90]. In another study, Li et al., have mentioned two lncRNAs, namely MAGI2-AS3 and CCDC144NL-AS1, derived from serum exosomes that encourage cellular proliferation, migration, and invasion in OC, via regulating the PI3K-AKT-mTOR pathway [91]. Several circRNAs have also been found to have their role as potent biomarkers in OC. A high level of circ_0000199 was seen in circulating exosomes of OC patients, which was found to be associated with poor survival outcomes, proving itself as a potent biomarker for OC [92]. The circ_0001874 and circ_0001971 derived from the saliva of patients were up-regulated in OC and identified as a potential diagnostic biomarker [93]. Table 5 enlists the expression of some of the ncRNAs, which are derived from exosomes, blood, serum/ plasma, and saliva samples, and have significance as biomarkers.
Table 5. ncRNAs derived from exosomes, blood, serum, and saliva.
| ncRNA Name | Sample | Expression Level in OC vs. Normal Cells | References |
|---|---|---|---|
| miR-8485 | Chondrocyte-derived exosomes | Higher | [94] |
| miRNA-1307-5p | Salivary exosomes | Higher | [89] |
| miR-200c-3p | Serum exosomes | Higher | [95][96] |
| miR-143 and miR-221 | Plasma exosomes | Lower and higher, respectively | [97][98] |
| miR-21 | Plasma/seminal exosomes | Higher | [99] |
| miR-31-5p | Macrophage-derived exosomes | Higher | [100] |
| miR-24-3p | Salivary exosomes | Higher | [101] |
| miR-382-5p | Fibroblast-associated exosomes | Higher | [102] |
| miR-10b | Plasma | Higher | [103] |
| miR-29a-3p | Serum | Higher | [104] |
| miR-486-5p | Plasma, salivary exosomes | Higher (in Stage II) | [105] |
| miR-155 | Exosomes | Higher | [106] |
| miR-142-3p | Exosomes | Higher | [107] |
| miR 455-5p and miR153 | Blood plasma | Higher and lower, respectively | [108] |
| miR-200b-3p, miR-483-5p, miR-425-5p, miR-374b-5p, miR-191-5p, miR-let-7c, miR-29a, miR-103, miR-1234, miR-638, miR-572, miR-22, miR-29b, miR-24-3p, miR-223, miR-20a, miR-29c, miR-17, miR-196a | Blood | Higher | [88] |
| miR-187, miR-9, miR-223, and miR-29c |
Lower | ||
| miR-494 | Whole blood | Higher | [109] |
| miR-16 and let-7b | Serum | Higher | [110] |
| miR-34a-5p | Cancer-associated fibroblast-derived exosomes | Lower | [111] |
| miR-3651 | Whole blood | Lower | [112] |
| lncRNAs MAGI2-AS3 and CCDC144NL-AS1 | Serum exosomes | Higher | [91] |
| lncRNA TIRY | Cancer-associated fibroblast-derived exosomes | Higher | [90] |
| lncRNA ADAMTS9-AS2 | Saliva, exosomes | Lower | [113] |
| circ_0069313 | Exosomes | Higher | [114] |
| circ_0000199 | Serum exosomes | Higher | [92] |
| circ_0026611 | Serum exosomes | Higher | [115] |
| circ_0001874 | Salivary exosomes | Higher | [93] |
| circ_0001971 | Salivary exosomes | Higher | [93] |
6. Non-Coding RNA in Oral Cancer Clinical Trials
The utilization of cancer-specific ncRNAs as biomarkers and therapeutic targets could aid in tailoring treatment options to specific patients or patient subgroups. In contrast to the typical tumor markers being used in latest medical applications, such as protein biomarkers and metabolic products, growing research indicates that ncRNAs could be ideal agents in cancer diagnosis and therapy. This is because the ncRNAs target multiple druggable and non-druggable targets and signaling events at a time. Further, they have tissue specificity, distinct RNA attributes of rapid detection, extra tissue-associated activity, and a far more stable structure [116]. Several clinical trials have been conducted using ncRNAs, especially using miRNAs as diagnostic and therapeutic biomarkers [117]. Through head-on targeting of gene sequences using antisense oligonucleotides (ASOs) and siRNA-associated therapeutic applications, the most sophisticated and concise therapeutic efforts at RNA screening have been achieved to date [117]. The lncRNA H19 promoter sequence has been introduced with the coding sequence of diphtheria toxin in BC-819 plasmid in clinical trials of the bladder, pancreatic and ovarian cancer [29][118]. The lncRNA HOTAIR and CCAT1 (ClinicalTrials.gov) are in clinical trials for thyroid and colorectal cancer diagnostic biomarker studies, whereas inhibitors of LINC01212 and lncMyoD are used as therapeutic markers for melanoma (US2016271163) and sarcoma therapy (WO2015020960) [29][118]. All these studies indicate possible clinical implications of ncRNAs in cancer diagnosis and therapy. A clinical study has been initiated to evaluate the sensitivity and specificity of miRNA-412 and miR-512 in extracellular vesicles from saliva in the malignant progression of OC (ClinicalTrials.gov Identifier: NCT04913545). Another trial has recently been initiated to identify diagnostic and prognostic miRNA biomarkers from blood, saliva, and tissue samples of head and neck cancer (ClinicalTrials.gov Identifier: NCT04305366). The diagnostic and therapeutic importance of lncRNA MALAT1 and its target miR-124 has been studied in saliva samples from OC patients (ClinicalTrials.gov Identifier: NCT05708209). A randomized phase II study has been initiated to identify salivary and plasma miRNAs of head and neck cancer patients, and monitor their change during the dietary intervention (ClinicalTrials.gov Identifier: NCT02869399). Two studies have been performed to assess the correlation of head–neck cancer immunotherapy with blood and plasma miRNAs profiles (ClinicalTrials.gov Identifier: NCT03843515 and NCT04453046). Low EGFR-AS1 lncRNA expression is determined as companion diagnostic biomarker of OC, and a phase II clinical study is recently ongoing to evaluate therapeutic efficacy of a certain drug of EGFR-associated advanced OC with low-EGFR-AS1 OC patients (ClinicalTrials.gov Identifier: NCT04946968). Outcomes of the study may indicate the diagnostic and therapeutic importance of EGFR-AS1 in OC treatment.
This entry is adapted from the peer-reviewed paper 10.3390/cancers15153752
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48.
- Kumar, M.; Nanavati, R.; Modi, T.G.; Dobariya, C. Oral cancer: Etiology and risk factors: A review. J. Cancer Res. Ther. 2016, 12, 458–463.
- Wong, T.; Wiesenfeld, D. Oral Cancer. Aust. Dent. J. 2018, 63 (Suppl. S1), S91–S99.
- Sujir, N.; Ahmed, J.; Pai, K.; Denny, C.; Shenoy, N. Challenges in Early Diagnosis of Oral Cancer: Cases Series. Acta Stomatol. Croat. 2019, 53, 174–180.
- Ai, L.; Chen, J.; Yan, H.; He, Q.; Luo, P.; Xu, Z.; Yang, X. Research Status and Outlook of PD-1/PD-L1 Inhibitors for Cancer Therapy. Drug Des. Dev. Ther. 2020, 14, 3625–3649.
- Gharat, S.A.; Momin, M.; Bhavsar, C. Oral Squamous Cell Carcinoma: Current Treatment Strategies and Nanotechnology-Based Approaches for Prevention and Therapy. Crit. Rev. Ther. Drug Carr. Syst. 2016, 33, 363–400.
- Dhamija, S.; Menon, M.B. Non-coding transcript variants of protein-coding genes—What are they good for? RNA Biol. 2018, 15, 1025–1031.
- Le, P.; Romano, G.; Nana-Sinkam, P.; Acunzo, M. Non-Coding RNAs in Cancer Diagnosis and Therapy: Focus on Lung Cancer. Cancers 2021, 13, 1372.
- Baxter, D.E.; Allinson, L.M.; Al Amri, W.S.; Poulter, J.A.; Pramanik, A.; Thorne, J.L.; Verghese, E.T.; Hughes, T.A. MiR-195 and Its Target SEMA6D Regulate Chemoresponse in Breast Cancer. Cancers 2021, 13, 5979.
- Chaurasia, A.; Alam, S.I.; Singh, N. Oral cancer diagnostics: An overview. Natl. J. Maxillofac. Surg. 2021, 12, 324–332.
- Basheeth, N.; Patil, N. Biomarkers in Head and Neck Cancer an Update. Indian J. Otolaryngol. Head Neck Surg. 2019, 71, 1002–1011.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Fang, C.; Li, Y. Prospective applications of microRNAs in oral cancer. Oncol. Lett. 2019, 18, 3974–3984.
- Jung, H.M.; Phillips, B.L.; Patel, R.S.; Cohen, D.M.; Jakymiw, A.; Kong, W.W.; Cheng, J.Q.; Chan, E.K. Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. J. Biol. Chem. 2012, 287, 29261–29272.
- Kawakita, A.; Yanamoto, S.; Yamada, S.; Naruse, T.; Takahashi, H.; Kawasaki, G.; Umeda, M. MicroRNA-21 promotes oral cancer invasion via the Wnt/beta-catenin pathway by targeting DKK2. Pathol. Oncol. Res. 2014, 20, 253–261.
- Zheng, G.; Li, N.; Jia, X.; Peng, C.; Luo, L.; Deng, Y.; Yin, J.; Song, Y.; Liu, H.; Lu, M.; et al. MYCN-mediated miR-21 overexpression enhances chemo-resistance via targeting CADM1 in tongue cancer. J. Mol. Med. 2016, 94, 1129–1141.
- Feng, Y.H.; Tsao, C.J. Emerging role of microRNA-21 in cancer. Biomed. Rep. 2016, 5, 395–402.
- Doukas, S.G.; Vageli, D.P.; Lazopoulos, G.; Spandidos, D.A.; Sasaki, C.T.; Tsatsakis, A. The Effect of NNK, A Tobacco Smoke Carcinogen, on the miRNA and Mismatch DNA Repair Expression Profiles in Lung and Head and Neck Squamous Cancer Cells. Cells 2020, 9, 1031.
- Kirave, P.; Gondaliya, P.; Kulkarni, B.; Rawal, R.; Garg, R.; Jain, A.; Kalia, K. Exosome mediated miR-155 delivery confers cisplatin chemoresistance in oral cancer cells via epithelial-mesenchymal transition. Oncotarget 2020, 11, 1157–1171.
- Ni, Y.H.; Huang, X.F.; Wang, Z.Y.; Han, W.; Deng, R.Z.; Mou, Y.B.; Ding, L.; Hou, Y.Y.; Hu, Q.G. Upregulation of a potential prognostic biomarker, miR-155, enhances cell proliferation in patients with oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 117, 227–233.
- Yete, S.; Saranath, D. MicroRNAs in oral cancer: Biomarkers with clinical potential. Oral Oncol. 2020, 110, 105002.
- Kong, W.; Yang, H.; He, L.; Zhao, J.J.; Coppola, D.; Dalton, W.S.; Cheng, J.Q. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 2008, 28, 6773–6784.
- Pang, X.; Tang, Y.L.; Liang, X.H. Transforming growth factor-beta signaling in head and neck squamous cell carcinoma: Insights into cellular responses. Oncol. Lett. 2018, 16, 4799–4806.
- Su, J.L.; Chen, P.S.; Johansson, G.; Kuo, M.L. Function and regulation of let-7 family microRNAs. MicroRNA 2012, 1, 34–39.
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415.
- Chang, C.J.; Hsu, C.C.; Chang, C.H.; Tsai, L.L.; Chang, Y.C.; Lu, S.W.; Yu, C.H.; Huang, H.S.; Wang, J.J.; Tsai, C.H.; et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol. Rep. 2011, 26, 1003–1010.
- Hunt, S.; Jones, A.V.; Hinsley, E.E.; Whawell, S.A.; Lambert, D.W. MicroRNA-124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett. 2011, 585, 187–192.
- Sur, S.; Ray, R.B. Emerging role of lncRNA ELDR in development and cancer. FEBS J. 2022, 289, 3011–3023.
- Gao, N.; Li, Y.; Li, J.; Gao, Z.; Yang, Z.; Li, Y.; Liu, H.; Fan, T. Long Non-Coding RNAs: The Regulatory Mechanisms, Research Strategies, and Future Directions in Cancers. Front. Oncol. 2020, 10, 598817.
- Huang, W.; Li, H.; Yu, Q.; Xiao, W.; Wang, D.O. LncRNA-mediated DNA methylation: An emerging mechanism in cancer and beyond. J. Exp. Clin. Cancer Res. CR 2022, 41, 100.
- Ishteyaq Majeed, S.; Mashooq Ahmad, D.; Kaiser Ahmad, B.; Tashook Ahmad, D.; Fayaz, A.; Syed Mudasir, A. Long Non-Coding RNAs: Biogenesis, Mechanism of Action and Role in Different Biological and Pathological Processes. In Recent Advances in Noncoding RNAs; Lütfi, T., Ed.; IntechOpen: Rijeka, Croatia, 2022; p. 118.
- Aznaourova, M.; Schmerer, N.; Schmeck, B.; Schulte, L.N. Disease-Causing Mutations and Rearrangements in Long Non-coding RNA Gene Loci. Front. Genet. 2020, 11, 527484.
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92.
- Xu, Y.; Jiang, E.; Shao, Z.; Shang, Z. Long Noncoding RNAs in the Metastasis of Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 616717.
- Ye, D.; Deng, Y.; Shen, Z. The Role and Mechanism of MALAT1 Long Non-Coding RNA in the Diagnosis and Treatment of Head and Neck Squamous Cell Carcinoma. OncoTargets Ther. 2021, 14, 4127–4136.
- Fu, S.; Wang, Y.; Li, H.; Chen, L.; Liu, Q. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag. Res. 2020, 12, 10181–10198.
- Sur, S.; Nakanishi, H.; Steele, R.; Zhang, D.; Varvares, M.A.; Ray, R.B. Long non-coding RNA ELDR enhances oral cancer growth by promoting ILF3-cyclin E1 signaling. EMBO Rep. 2020, 21, e51042.
- Sur, S.; Steele, R.; Ko, B.C.B.; Zhang, J.; Ray, R.B. Long noncoding RNA ELDR promotes cell cycle progression in normal oral keratinocytes through induction of a CTCF-FOXM1-AURKA signaling axis. J. Biol. Chem. 2022, 298, 101895.
- Zeng, B.; Li, Y.; Jiang, F.; Wei, C.; Chen, G.; Zhang, W.; Zhao, W.; Yu, D. LncRNA GAS5 suppresses proliferation, migration, invasion, and epithelial-mesenchymal transition in oral squamous cell carcinoma by regulating the miR-21/PTEN axis. Exp. Cell Res. 2019, 374, 365–373.
- Chen, P.Y.; Hsieh, P.L.; Peng, C.Y.; Liao, Y.W.; Yu, C.H.; Yu, C.C. LncRNA MEG3 inhibits self-renewal and invasion abilities of oral cancer stem cells by sponging miR-421. J. Formos. Med. Assoc. = Taiwan Yi Zhi 2021, 120, 1137–1142.
- Zhang, L.; Meng, X.; Zhu, X.W.; Yang, D.C.; Chen, R.; Jiang, Y.; Xu, T. Long non-coding RNAs in Oral squamous cell carcinoma: Biologic function, mechanisms and clinical implications. Mol. Cancer 2019, 18, 102.
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53.
- Andresini, O.; Ciotti, A.; Rossi, M.N.; Battistelli, C.; Carbone, M.; Maione, R. A cross-talk between DNA methylation and H3 lysine 9 dimethylation at the KvDMR1 region controls the induction of Cdkn1c in muscle cells. Epigenetics 2016, 11, 791–803.
- Wang, M.; Yu, F.; Wu, W.; Zhang, Y.; Chang, W.; Ponnusamy, M.; Wang, K.; Li, P. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 2017, 13, 1497–1506.
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461.
- Chen, L.L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388.
- Geng, X.; Jia, Y.; Zhang, Y.; Shi, L.; Li, Q.; Zang, A.; Wang, H. Circular RNA: Biogenesis, degradation, functions and potential roles in mediating resistance to anticarcinogens. Epigenomics 2020, 12, 267–283.
- Yu, T.; Wang, Y.; Fan, Y.; Fang, N.; Wang, T.; Xu, T.; Shu, Y. CircRNAs in cancer metabolism: A review. J. Hematol. Oncol. 2019, 12, 90.
- Zeng, Y.; Zou, Y.; Gao, G.; Zheng, S.; Wu, S.; Xie, X.; Tang, H. The biogenesis, function and clinical significance of circular RNAs in breast cancer. Cancer Biol. Med. 2021, 19, 14–29.
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14.
- Fan, H.Y.; Jiang, J.; Tang, Y.J.; Liang, X.H.; Tang, Y.L. CircRNAs: A New Chapter in Oral Squamous Cell Carcinoma Biology. OncoTargets Ther. 2020, 13, 9071–9083.
- Chen, X.; Yu, J.; Tian, H.; Shan, Z.; Liu, W.; Pan, Z.; Ren, J. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J. Cell. Physiol. 2019, 234, 19130–19140.
- He, T.; Li, X.; Xie, D.; Tian, L. Overexpressed circPVT1 in oral squamous cell carcinoma promotes proliferation by serving as a miRNA sponge. Mol. Med. Rep. 2019, 20, 3509–3518.
- Gao, L.; Dou, Z.C.; Ren, W.H.; Li, S.M.; Liang, X.; Zhi, K.Q. CircCDR1as upregulates autophagy under hypoxia to promote tumor cell survival via AKT/ERK((1/2))/mTOR signaling pathways in oral squamous cell carcinomas. Cell Death Dis. 2019, 10, 745.
- Su, W.; Wang, Y.; Wang, F.; Zhang, B.; Zhang, H.; Shen, Y.; Yang, H. Circular RNA hsa_circ_0007059 indicates prognosis and influences malignant behavior via AKT/mTOR in oral squamous cell carcinoma. J. Cell. Physiol. 2019, 234, 15156–15166.
- Su, W.; Wang, Y.; Wang, F.; Sun, S.; Li, M.; Shen, Y.; Yang, H. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer 2019, 19, 400.
- Papatsirou, M.; Artemaki, P.I.; Karousi, P.; Scorilas, A.; Kontos, C.K. Circular RNAs: Emerging Regulators of the Major Signaling Pathways Involved in Cancer Progression. Cancers 2021, 13, 2744.
- Gao, L.; Zhao, C.; Li, S.; Dou, Z.; Wang, Q.; Liu, J.; Ren, W.; Zhi, K. circ-PKD2 inhibits carcinogenesis via the miR-204-3p/APC2 axis in oral squamous cell carcinoma. Mol. Carcinog. 2019, 58, 1783–1794.
- Dragon, F.; Lemay, V.; Trahan, C. snoRNAs: Biogenesis, Structure and Function. In Encyclopedia of Life Sciences; Wiley: Hoboken, NJ, USA, 2006.
- Chamorro-Petronacci, C.; Perez-Sayans, M.; Padin-Iruegas, M.E.; Marichalar-Mendia, X.; Gallas-Torreira, M.; Garcia Garcia, A. Differential expression of snoRNAs in oral squamous cell carcinomas: New potential diagnostic markers. J. Enzym. Inhib. Med. Chem. 2018, 33, 424–427.
- Xing, L.; Zhang, X.; Zhang, X.; Tong, D. Expression scoring of a small-nucleolar-RNA signature identified by machine learning serves as a prognostic predictor for head and neck cancer. J. Cell. Physiol. 2020, 235, 8071–8084.
- Liu, Z.; Tao, H. Small nucleolar RNA host gene 3 facilitates cell proliferation and migration in oral squamous cell carcinoma via targeting nuclear transcription factor Y subunit gamma. J. Cell. Biochem. 2020, 121, 2150–2158.
- Lu, N.; Yin, Y.; Yao, Y.; Zhang, P. SNHG3/miR-2682-5p/HOXB8 promotes cell proliferation and migration in oral squamous cell carcinoma. Oral Dis. 2021, 27, 1161–1170.
- Wang, T.; Liang, D.; Yang, H. SNHG15 facilitated malignant behaviors of oral squamous cell carcinoma through targeting miR-188-5p/DAAM1. J. Oral Pathol. Med. 2021, 50, 681–691.
- Liu, P.; Dong, Y.; Gu, J.; Puthiyakunnon, S.; Wu, Y.; Chen, X.G. Developmental piRNA profiles of the invasive vector mosquito Aedes albopictus. Parasites Vectors 2016, 9, 524.
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006, 442, 199–202.
- Han, Y.N.; Li, Y.; Xia, S.Q.; Zhang, Y.Y.; Zheng, J.H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem. 2017, 44, 1–20.
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9.
- Reeves, M.E.; Firek, M.; Jliedi, A.; Amaar, Y.G. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget 2017, 8, 34268–34282.
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123.
- Wu, L.; Jiang, Y.; Zheng, Z.; Li, H.; Cai, M.; Pathak, J.L.; Li, Z.; Huang, L.; Zeng, M.; Zheng, H.; et al. mRNA and P-element-induced wimpy testis-interacting RNA profile in chemical-induced oral squamous cell carcinoma mice model. Exp. Anim. 2020, 69, 168–177.
- Saad, M.A.; Ku, J.; Kuo, S.Z.; Li, P.X.; Zheng, H.; Yu, M.A.; Wang-Rodriguez, J.; Ongkeko, W.M. Identification and characterization of dysregulated P-element induced wimpy testis-interacting RNAs in head and neck squamous cell carcinoma. Oncol. Lett. 2019, 17, 2615–2622.
- Firmino, N.; Martinez, V.D.; Rowbotham, D.A.; Enfield, K.S.S.; Bennewith, K.L.; Lam, W.L. HPV status is associated with altered PIWI-interacting RNA expression pattern in head and neck cancer. Oral Oncol. 2016, 55, 43–48.
- Krishnan, A.R.; Qu, Y.; Li, P.X.; Zou, A.E.; Califano, J.A.; Wang-Rodriguez, J.; Ongkeko, W.M. Computational methods reveal novel functionalities of PIWI-interacting RNAs in human papillomavirus-induced head and neck squamous cell carcinoma. Oncotarget 2018, 9, 4614–4624.
- Chattopadhyay, T.; Gupta, P.; Nayak, R.; Mallick, B. Genome-wide profiling of dysregulated piRNAs and their target genes implicated in oncogenicity of tongue squamous cell carcinoma. Gene 2023, 849, 146919.
- Krishnan, A.R.; Korrapati, A.; Zou, A.E.; Qu, Y.; Wang, X.Q.; Califano, J.A.; Wang-Rodriguez, J.; Lippman, S.M.; Hovell, M.F.; Ongkeko, W.M. Smoking status regulates a novel panel of PIWI-interacting RNAs in head and neck squamous cell carcinoma. Oral Oncol. 2017, 65, 68–75.
- Celetti, A.; Merolla, F.; Luise, C.; Siano, M.; Staibano, S. Novel Markers for Diagnosis and Prognosis of Oral Intraepithelial Neoplasia; IntechOpen: Rijeka, Croatia, 2012.
- Chattopadhyay, E.; Singh, R.; Ray, A.; Roy, R.; De Sarkar, N.; Paul, R.R.; Pal, M.; Aich, R.; Roy, B. Expression deregulation of mir31 and CXCL12 in two types of oral precancers and cancer: Importance in progression of precancer and cancer. Sci. Rep. 2016, 6, 32735.
- Osan, C.; Chira, S.; Nutu, A.M.; Braicu, C.; Baciut, M.; Korban, S.S.; Berindan-Neagoe, I. The Connection between MicroRNAs and Oral Cancer Pathogenesis: Emerging Biomarkers in Oral Cancer Management. Genes 2021, 12, 1989.
- Grubelnik, G.; Bostjancic, E.; Anicin, A.; Dovsak, T.; Zidar, N. MicroRNAs and Long Non-Coding RNAs as Regulators of NANOG Expression in the Development of Oral Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 579053.
- Zhang, K.; Qiu, W.; Wu, B.; Fang, F. Long non-coding RNAs are novel players in oral inflammatory disorders, potentially premalignant oral epithelial lesions and oral squamous cell carcinoma (Review). Int. J. Mol. Med. 2020, 46, 535–545.
- Jia, H.; Wang, X.; Sun, Z. Exploring the long noncoding RNAs-based biomarkers and pathogenesis of malignant transformation from dysplasia to oral squamous cell carcinoma by bioinformatics method. Eur. J. Cancer Prev. 2020, 29, 174–181.
- Xu, S.; Song, Y.; Shao, Y.; Zhou, H. Comprehensive analysis of circular RNA in oral leukoplakia: Upregulated circHLA-C as a potential biomarker for diagnosis and prognosis. Ann. Transl. Med. 2020, 8, 1375.
- Chen, C.M.; Chu, T.H.; Chou, C.C.; Chien, C.Y.; Wang, J.S.; Huang, C.C. Exosome-derived microRNAs in oral squamous cell carcinomas impact disease prognosis. Oral Oncol. 2021, 120, 105402.
- Momen-Heravi, F.; Bala, S. Extracellular vesicles in oral squamous carcinoma carry oncogenic miRNA profile and reprogram monocytes via NF-kappaB pathway. Oncotarget 2018, 9, 34838–34854.
- Yap, T.; Pruthi, N.; Seers, C.; Belobrov, S.; McCullough, M.; Celentano, A. Extracellular Vesicles in Oral Squamous Cell Carcinoma and Oral Potentially Malignant Disorders: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 1197.
- Patil, S.; Warnakulasuriya, S. Blood-based circulating microRNAs as potential biomarkers for predicting the prognosis of head and neck cancer-a systematic review. Clin. Oral Investig. 2020, 24, 3833–3841.
- Patel, A.; Patel, S.; Patel, P.; Mandlik, D.; Patel, K.; Tanavde, V. Salivary Exosomal miRNA-1307-5p Predicts Disease Aggressiveness and Poor Prognosis in Oral Squamous Cell Carcinoma Patients. Int. J. Mol. Sci. 2022, 23, 10639.
- Jin, N.; Jin, N.; Bu, W.; Li, X.; Liu, L.; Wang, Z.; Tong, J.; Li, D. Long non-coding RNA TIRY promotes tumor metastasis by enhancing epithelial-to-mesenchymal transition in oral cancer. Exp. Biol. Med. 2020, 245, 585–596.
- Li, C.; Guo, H.; Xiong, J.; Feng, B.; Zhu, P.; Jiang, W.; Jiang, P.; Su, X.; Huang, X. Exosomal long noncoding RNAs MAGI2-AS3 and CCDC144NL-AS1 in oral squamous cell carcinoma development via the PI3K-AKT-mTOR signaling pathway. Pathol. Res. Pract. 2022, 240, 154219.
- Luo, Y.; Liu, F.; Guo, J.; Gui, R. Upregulation of circ_0000199 in circulating exosomes is associated with survival outcome in OSCC. Sci. Rep. 2020, 10, 13739.
- Zhao, S.Y.; Wang, J.; Ouyang, S.B.; Huang, Z.K.; Liao, L. Salivary Circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as Novel Biomarkers for the Diagnosis of Oral Squamous Cell Carcinoma. Cell. Physiol. Biochem. 2018, 47, 2511–2521.
- Li, W.; Han, Y.; Zhao, Z.; Ji, X.; Wang, X.; Jin, J.; Wang, Q.; Guo, X.; Cheng, Z.; Lu, M.; et al. Oral mucosal mesenchymal stem cell-derived exosomes: A potential therapeutic target in oral premalignant lesions. Int. J. Oncol. 2019, 54, 1567–1578.
- Cui, J.; Wang, H.; Zhang, X.; Sun, X.; Zhang, J.; Ma, J. Exosomal miR-200c suppresses chemoresistance of docetaxel in tongue squamous cell carcinoma by suppressing TUBB3 and PPP2R1B. Aging 2020, 12, 6756–6773.
- Kawakubo-Yasukochi, T.; Morioka, M.; Hazekawa, M.; Yasukochi, A.; Nishinakagawa, T.; Ono, K.; Kawano, S.; Nakamura, S.; Nakashima, M. miR-200c-3p spreads invasive capacity in human oral squamous cell carcinoma microenvironment. Mol. Carcinog. 2018, 57, 295–302.
- He, L.; Liao, L.; Du, L. miR-144-3p inhibits tumor cell growth and invasion in oral squamous cell carcinoma through the downregulation of the oncogenic gene, EZH2. Int. J. Mol. Med. 2020, 46, 828–838.
- Husna, A.A.; Rahman, M.M.; Lai, Y.C.; Chen, H.W.; Hasan, M.N.; Nakagawa, T.; Miura, N. Identification of melanoma-specific exosomal miRNAs as the potential biomarker for canine oral melanoma. Pigment Cell Melanoma Res. 2021, 34, 1062–1073.
- Liu, T.; Chen, G.; Sun, D.; Lei, M.; Li, Y.; Zhou, C.; Li, X.; Xue, W.; Wang, H.; Liu, C.; et al. Exosomes containing miR-21 transfer the characteristic of cisplatin resistance by targeting PTEN and PDCD4 in oral squamous cell carcinoma. Acta Biochim. Et Biophys. Sin. 2017, 49, 808–816.
- Kavitha, M.; Jayachandran, D.; Aishwarya, S.Y.; Md Younus, P.; Venugopal, A.; Suresh Babu, H.W.; Ajay, E.; Sanjana, M.; Arul, N.; Balachandar, V. A new insight into the diverse facets of microRNA-31 in oral squamous cell carcinoma. Egypt. J. Med. Hum. Genet. 2022, 23, 149.
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2020, 121, 109553.
- Sun, L.P.; Xu, K.; Cui, J.; Yuan, D.Y.; Zou, B.; Li, J.; Liu, J.L.; Li, K.Y.; Meng, Z.; Zhang, B. Cancer-associated fibroblast-derived exosomal miR-382-5p promotes the migration and invasion of oral squamous cell carcinoma. Oncol. Rep. 2019, 42, 1319–1328.
- Lu, Y.C.; Chen, Y.J.; Wang, H.M.; Tsai, C.Y.; Chen, W.H.; Huang, Y.C.; Fan, K.H.; Tsai, C.N.; Huang, S.F.; Kang, C.J.; et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev. Res. 2012, 5, 665–674.
- Karimi, A.; Bahrami, N.; Sayedyahossein, A.; Derakhshan, S. Evaluation of circulating serum 3 types of microRNA as biomarkers of oral squamous cell carcinoma; A pilot study. J. Oral Pathol. Med. 2020, 49, 43–48.
- Faur, C.I.; Roman, R.C.; Jurj, A.; Raduly, L.; Almasan, O.; Rotaru, H.; Chirila, M.; Moldovan, M.A.; Hedesiu, M.; Dinu, C. Salivary Exosomal MicroRNA-486-5p and MicroRNA-10b-5p in Oral and Oropharyngeal Squamous Cell Carcinoma. Medicina 2022, 58, 1478.
- Sayyed, A.A.; Gondaliya, P.; Mali, M.; Pawar, A.; Bhat, P.; Khairnar, A.; Arya, N.; Kalia, K. MiR-155 Inhibitor-Laden Exosomes Reverse Resistance to Cisplatin in a 3D Tumor Spheroid and Xenograft Model of Oral Cancer. Mol. Pharm. 2021, 18, 3010–3025.
- Dickman, C.T.; Lawson, J.; Jabalee, J.; MacLellan, S.A.; LePard, N.E.; Bennewith, K.L.; Garnis, C. Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget 2017, 8, 15252–15266.
- Baber, S.; Bayat, M.; Mohamadnia, A.; Shamshiri, A.; Amini Shakib, P.; Bahrami, N. Role of miR153 and miR455-5p Expression in Oral Squamous Cell Carcinoma Isolated from Plasma. Asian Pac. J. Cancer Prev. 2021, 22, 157–161.
- Ries, J.; Vairaktaris, E.; Agaimy, A.; Kintopp, R.; Baran, C.; Neukam, F.W.; Nkenke, E. miR-186, miR-3651 and miR-494: Potential biomarkers for oral squamous cell carcinoma extracted from whole blood. Oncol. Rep. 2014, 31, 1429–1436.
- Maclellan, S.A.; Lawson, J.; Baik, J.; Guillaud, M.; Poh, C.F.; Garnis, C. Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med. 2012, 1, 268–274.
- Li, Y.Y.; Tao, Y.W.; Gao, S.; Li, P.; Zheng, J.M.; Zhang, S.E.; Liang, J.; Zhang, Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine 2018, 36, 209–220.
- Ries, J.; Baran, C.; Wehrhan, F.; Weber, M.; Motel, C.; Kesting, M.; Nkenke, E. The altered expression levels of miR-186, miR-494 and miR-3651 in OSCC tissue vary from those of the whole blood of OSCC patients. Cancer Biomark. 2019, 24, 19–30.
- Zhou, S.; Zhu, Y.; Li, Z.; Zhu, Y.; He, Z.; Zhang, C. Exosome-derived long non-coding RNA ADAMTS9-AS2 suppresses progression of oral submucous fibrosis via AKT signalling pathway. J. Cell. Mol. Med. 2021, 25, 2262–2273.
- Chen, Y.; Li, Z.; Liang, J.; Liu, J.; Hao, J.; Wan, Q.; Liu, J.; Luo, C.; Lu, Z. CircRNA has_circ_0069313 induced OSCC immunity escape by miR-325-3p-Foxp3 axes in both OSCC cells and Treg cells. Aging 2022, 14, 4376–4389.
- Ye, D.; Gong, M.; Deng, Y.; Fang, S.; Cao, Y.; Xiang, Y.; Shen, Z. Roles and clinical application of exosomal circRNAs in the diagnosis and treatment of malignant tumors. J. Transl. Med. 2022, 20, 161.
- Mazumder, S.; Datta, S.; Ray, J.G.; Chaudhuri, K.; Chatterjee, R. Liquid biopsy: miRNA as a potential biomarker in oral cancer. Cancer Epidemiol. 2019, 58, 137–145.
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nature reviews. Drug Discov. 2021, 20, 629–651.
- Qian, Y.; Shi, L.; Luo, Z. Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 2020, 7, 612393.
This entry is offline, you can click here to edit this entry!
