β-lactamase enzymes have generated significant interest due to their ability to confer resistance to the most commonly used family of antibiotics in human medicine. Among these enzymes, the class B β-lactamases are members of a superfamily of metallo-β-lactamase (MβL) fold proteins which are characterised by conserved motifs (i.e., HxHxDH) and are not only limited to bacteria.
- metallo-β-lactamase (MβL) fold proteins
- multifunctional enzymes
- antibiotic-hydrolysing activity
1. Introduction
2. Diversity of the Superfamily of Metallo-β-Lactamase (MβL) Fold Enzymes
3. MβL Fold Enzymes in Bacteria: Class B β-Lactamases
3.1. Distribution and Diversity of MβL Fold Enzymes in Bacteria
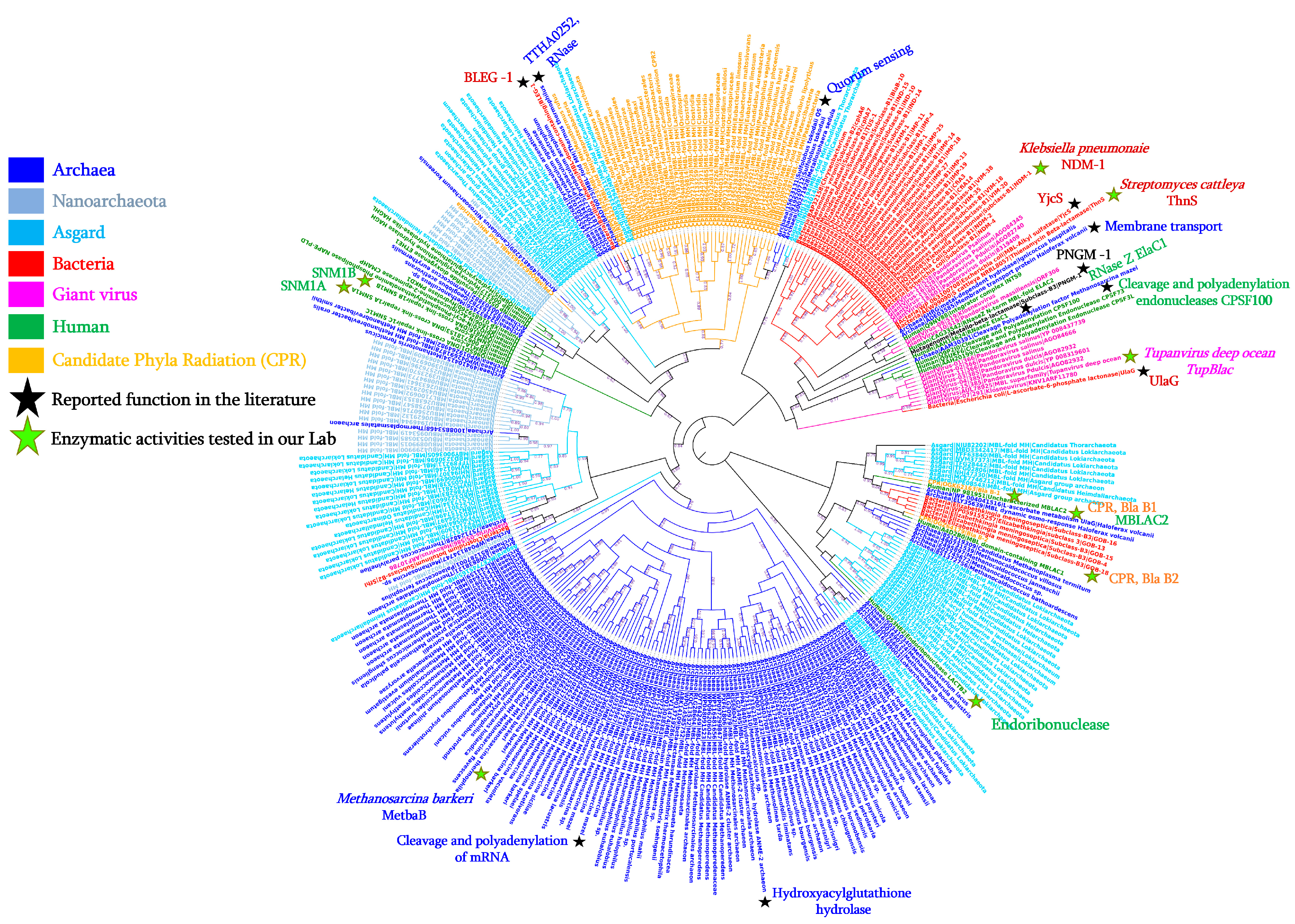
3.2. Reported Activities of Bacterial MβL Enzymes Other Than β-Lactams Hydrolysis
|
Kingdom |
Acc. Number |
Species |
Gene Name |
Size (aa) |
Default Annotation |
Reported Activities |
References |
|---|---|---|---|---|---|---|---|
|
Bacteria |
AGM20433 |
Enterobacter cloacae |
IMP-1 |
247 |
Imipenem hydrolysing β-lactamase |
β-lactamase; Ribonuclease |
[28] |
|
AEW99090 |
Streptomyces cattleya |
ThnS |
330 |
Putative β-lactamase |
Imipenemase; Ascorbic acid degradation; Nuclease; Ribonuclease |
[29] |
|
|
HQ328085 |
Klebsiella pneumoniae |
NDM-1 |
270 |
New Delhi metallo-β-lactamase |
β-lactamase; Ribonuclease |
In this study |
|
|
NA |
Streptococcus pneumoniae |
- |
363 |
L-ascorbate 6-phosphate lactonase |
β-lactamase |
[26] |
|
|
P39300.2 |
Escherichia coli |
UlaG |
354 |
L-ascorbate-6-phosphate lactonase |
β-lactamase; Ribonuclease |
[27] |
|
|
WP_010974862 |
Agrobacterium tumefaciens |
AiiB |
276 |
Zn-dependent hydrolases |
Quorum-quenching lactonase |
[32] |
|
|
PDB: 7EV5_A |
Bacillus lehensis |
BLEG-1 |
210 |
β-lactamase domain containing protein |
β-lactamase; Glyoxalase II |
[30] |
|
|
NA |
Soil metagenome |
MβLp01 |
312 |
Metallo-β-lactamase fold protein |
β-lactamase; Phytase |
[33] |
|
|
NA |
Soil metagenome |
MβLP02 |
355 |
Metallo-β-lactamase fold protein |
β-lactamase; Phytase |
[33] |
|
|
PDB: 7BZ4_B |
Deep-seep sediments metagenome |
PNGM-1 |
372 |
Metallo-β-lactamase |
β-lactamase; Ribonuclease |
[34] |
|
|
WP_063100708 |
Escherichia coli |
YjcS |
661 |
Uncharacterised protein |
Alkyl sulfatase |
[35] |
|
|
Archaea |
ELY35639 |
Haloferax volcanii |
HVO_2763 |
278 |
Zn-dependent hydrolases of the β-lactamase fold |
Membrane transport; Static and dynamic osmo-response |
[36] |
|
851225341 |
Methanosarcina barkeri |
MetbaB |
214 |
MβL fold metallo-hydrolase |
β-lactamase; Ribonuclease; and D-lactate hydrolase |
[37] |
|
|
15623131 |
Sulfolobus tokodaii |
- |
200 |
Putative hydrolase |
Quorum sensing activity |
[38] |
|
|
AAM30391 |
Methanosarcina mazei |
- |
638 |
Metal-dependent RNase, contains metallo-β-lactamase and KH domains |
Cleavage and Polyadenylation of mRNA |
[39] |
|
|
BAD70075 |
Thermus thermophilus |
TTHA0252 |
Metallo-β-lactamase superfamily |
Ribonuclease (RNase) |
[40] |
||
|
NA |
Methanocaldococcus jannaschii |
MjRNase J1 |
Ribonuclease Rnase J |
Ribonuclease (RNase) |
[41] |
||
|
NA |
Methanocaldococcus jannaschii |
MjRNase J2 |
Ribonuclease Rnase J |
Ribonuclease (RNase); Nuclease |
|||
|
NA |
Methanocaldococcus jannaschii |
MjRNase J3 |
Ribonuclease Rnase J |
Ribonuclease (RNase); Nuclease |
|||
|
Human |
NP_981951 |
Homo sapiens |
MβLAC2 |
280 |
Metallo-β-lactamase domain |
Exosome biogenesis enzyme; β-lactamase |
|
|
Q6PJP8 |
Homo sapiens |
SNM1A |
365 |
DNA cross-link repair 1A |
Cisplatin or Mitomycin hydrolase; β-lactamase |
||
|
Q9H816 |
Homo sapiens |
SNM1B |
335 |
5′ exonuclease Apollo isoform X1 |
β-lactamase |
[43] |
|
|
Giant viruses |
AUL78925 |
Tupanvirus deep ocean |
TupBlac |
322 |
β-lactamase superfamily domain |
β-lactamase; Ribonuclease |
[45] |
|
Candidate Phyla Radiation (CPR) |
KKR15801 |
Candidatus Levybacteria |
- |
287 |
β-lactamase class A-like protein |
β-lactamase; Ribonuclease |
[46] |
|
KKR17584 |
Candidatus Levybacteria |
- |
303 |
β-lactamase class A-like protein |
β-lactamase; Ribonuclease |
||
|
OGK62163 |
Candidatus Roizmanbacteria |
- |
263 |
Hypothetical protein |
β-lactamase; Ribonuclease |
||
|
OGZ64179 |
Candidatus Staskawiczbacteria |
- |
251 |
Hypothetical protein |
β-lactamase; Ribonuclease |
||
|
QHU90009 |
Candidatus Saccharibacteria |
- |
722 |
RNase J family beta-CASP ribonuclease |
β-lactamase; Ribonuclease |
This entry is adapted from the peer-reviewed paper 10.3390/cells12131752
References
- Jacoby George, A. β-Lactamase Nomenclature. Antimicrob. Agents Chemother. 2006, 50, 1123–1129.
- Garau, J. Beta-lactamases: Current situation and clinical importance. Intensive Care Med. 1994, 20, S5–S9.
- Caetano-Anollés, D.; Kim, K.M.; Mittenthal, J.E.; Caetano-Anollés, G. Proteome evolution and the metabolic origins of translation and cellular life. J. Mol. Evol. 2011, 72, 14–33.
- Risso, V.A.; Gavira, J.A.; Mejia-Carmona, D.F.; Gaucher, E.A.; Sanchez-Ruiz, J.M. Hyperstability and substrate promiscuity in laboratory resurrections of precambrian β-lactamases. J. Am. Chem. Soc. 2013, 135, 2899–2902.
- Thornton, J.W. Resurrecting ancient genes: Experimental analysis of extinct molecules. Nat. Rev. Genet. 2004, 5, 366–375.
- Bush, K. The ABCD’s of β-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559.
- Bush, K. Past and Present Perspectives on beta-lactamases. Antimicrob. Agents Chemother. 2018, 62, e01076-18.
- Bebrone, C. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 2007, 74, 1686–1701.
- Fröhlich, C.; Chen, J.Z.; Gholipour, S.; Erdogan, A.N.; Tokuriki, N. Evolution of β-lactamases and enzyme promiscuity. Protein Eng. Des. Sel. 2021, 34, gzab013.
- Marasinghe, G.P.K.; Sander, I.M.; Bennett, B.; Periyannan, G.; Yang, K.W.; Makaroff, C.A.; Crowder, M.W. Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 2005, 280, 40668–40675.
- Aravind, L. An evolutionary classification of the metallo-beta-lactamase fold proteins. Silico Biol. 1999, 1, 69–91.
- Baier, F.; Tokuriki, N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J. Mol. Biol. 2014, 426, 2442–2456.
- Bergonzi, C.; Schwab, M.; Naik, T.; Daudé, D.; Chabrière, E.; Elias, M. Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties. Sci. Rep. 2018, 8, 19–21.
- Perez-Garcia, P.; Kobus, S.; Gertzen, C.G.W.; Hoeppner, A.; Holzscheck, N.; Strunk, C.H.; Huber, H.; Jaeger, K.E.; Gohlke, H.; Kovacic, F.; et al. A promiscuous ancestral enzyme’s structure unveils protein variable regions of the highly diverse metallo-β-lactamase family. Commun. Biol. 2021, 4, 132.
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104.
- Laham, S.M.; Gonsalkorale, K.; Von Hippel, W. Darwinian grandparenting: Preferential investment in more certain kin. Personal. Soc. Psychol. Bull. 2005, 31, 63–72.
- Keshri, V.; Panda, A.; Levasseur, A.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. Phylogenomic Analysis of β-Lactamase in Archaea and Bacteria Enables the Identification of Putative New Members. Genome Biol. Evol. 2018, 10, 1106–1114.
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431.
- Atkinson, H.J.; Morris, J.H.; Ferrin, T.E.; Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 2009, 4, e4345.
- Somboro, A.M.; Sekyere, J.O.; Amoako, D.G.; Essack, S.Y.; Bester, L.A. Diversity and proliferation of metallo-β-lactamases: A clarion call for clinically effective metallo-β-lactamase inhibitors. Appl. Environ. Microbiol. 2018, 84, e00698-18.
- Salahuddin, P.; Kumar, A.; Khan, A.U. Structure, Function of Serine and Metallo-β-lactamases and their Inhibitors. Curr. Protein Pept. Sci. 2018, 19, 130–144.
- Heinz, U.; Adolph, H.W. Metallo-β-lactamases: Two binding sites for one catalytic metal ion? Cell. Mol. Life Sci. 2004, 61, 2827–2839.
- Sabath, L.D.; Abraham, E.P. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem. J. 1966, 98, 10–12.
- Carfi, A.; Pares, S.; Duée, E.; Galleni, M.; Duez, C.; Frère, J.M.; Dideberg, O. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995, 14, 4914–4921.
- Daiyasu, H.; Osaka, K.; Ishino, Y.; Toh, H. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 2001, 503, 1–6.
- Chang, C.Y.; Lin, H.J.; Li, B.R.; Li, Y.K. A Novel metallo-β-lactamase involved in the ampicillin resistance of Streptococcus pneumoniae ATCC 49136 strain. PLoS ONE 2016, 11, e0155905.
- Garces, F.; Fernández, F.J.; Montellà, C.; Penya-Soler, E.; Prohens, R.; Aguilar, J.; Baldomà, L.; Coll, M.; Badia, J.; Vega, M.C. Molecular architecture of the Mn2+-dependent lactonase UlaG reveals an RNase-like metallo-β-lactamase fold and a novel quaternary structure. J. Mol. Biol. 2010, 398, 715–729.
- Kato, Y.; Takahashi, M.; Seki, M.; Nashimoto, M.; Shimizu-Ibuka, A. RNA-hydrolyzing activity of metallo-βlactamase IMP-1. PLoS ONE 2020, 15, e0241557.
- Diene, S.M.; Pinault, L.; Baron, S.A.; Azza, S.; Armstrong, N.; Hadjadj, L.; Chabrière, E.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. A metallo-β-lactamase enzyme for internal detoxification of the antibiotic thienamycin. Sci. Rep. 2021, 11, 10062.
- Au, S.X.; Dzulkifly, N.S.; Noor, N.D.M.; Matsumura, H.; Rahman, R.N.Z.R.A.; Normi, Y.M. Dual activity bleg-1 from bacillus lehensis g1 revealed structural resemblance to b3 metallo-β-lactamase and glyoxalase ii: An insight into its enzyme promiscuity and evolutionary divergence. Int. J. Mol. Sci. 2021, 22, 9377.
- Tan, S.H.; Normi, Y.M.; Leow, A.T.C.; Salleh, A.B.; Murad, A.M.A.; Mahadi, N.M.; Abdul Rahman, M.B. Danger lurking in the “unknowns”: Structure-to-Function Studies of Hypothetical Protein Bleg1_2437 from Bacillus lehensis G1 Alkaliphile Revealed an Evolutionary Divergent B3 Metallo-beta-lactamase. J. Biochem. Adv. 2016, 161, 167–186.
- Liu, D.; Thomas, P.W.; Momb, J.; Hoang, Q.Q.; Petsko, G.A.; Ringe, D.; Fast, W. Structure and Specificity of a Quorum-Quenching Lactonase ( AiiB ) from. Biochemistry 2007, 46, 11789–11799.
- Castillo Villamizar, G.A.; Funkner, K.; Nacke, H.; Foerster, K.; Daniel, R. Functional Metagenomics Reveals a New Catalytic Domain, the Metallo-β-Lactamase Superfamily Domain, Associated with Phytase Activity. mSphere 2019, 4.
- Lee, J.H.; Takahashi, M.; Jeon, J.H.; Kang, L.-W.; Seki, M.; Park, K.S.; Hong, M.-K.; Park, Y.S.; Kim, T.Y.; Karim, A.M.; et al. Dual activity of PNGM-1, a metallo-β-lactamase and tRNase Z, pinpoints the evolutionary origin of subclass B3 metallo-β-lactamases. bioRxiv 2019, bioRxiv:575373.
- Liang, Y.; Gao, Z.; Dong, Y.; Liu, Q. Structural and functional analysis show that the Escherichia coli uncharacterized protein YjcS is likely an alkylsulfatase. Protein Sci. 2014, 23, 1442–1450.
- Fischer, S.; John von Freyend, S.; Sabag-Daigle, A.; Daniels, C.J.; Allers, T.; Marchfelder, A. Assigning a function to a conserved archaeal metallo-β-lactamase from Haloferax volcanii. Extremophiles 2012, 16, 333–343.
- Diene, S.M.; Pinault, L.; Armstrong, N.; Azza, S.; Keshri, V.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Rolain, J.M.; Pontarotti, P.; et al. Dual rnase and β-lactamase activity of a single enzyme encoded in archaea. Life 2020, 10, 280.
- Shimada, A.; Ishikawa, H.; Nakagawa, N.; Kuramitsu, S.; Masui, R. The first crystal structure of an archaeal metallo-β-lactamase superfamily protein; ST1585 from Sulfolobus tokodaii. Proteins Struct. Funct. Bioinform. 2010, 78, 2399–2402.
- Mir-Montazeri, B.; Ammelburg, M.; Forouzan, D.; Lupas, A.N.; Hartmann, M.D. Crystal structure of a dimeric archaeal Cleavage and Polyadenylation Specificity Factor. J. Struct. Biol. 2011, 173, 191–195.
- Ishikawa, H.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Crystal structure of TTHA0252 from Thermus thermophilus HB8, a RNA degradation protein of the metallo-??-lactamase superfamily. J. Biochem. 2006, 140, 535–542.
- Levy, S.; Portnoy, V.; Admon, J.; Schuster, G. Distinct activities of several RNase J proteins in methanogenic archaea. RNA Biol. 2011, 8, 1073–1083.
- Buschow, S.I.; van Balkom, B.W.M.; Aalberts, M.; Heck, A.J.R.; Wauben, M.; Stoorvogel, W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol. Cell Biol. 2010, 88, 851–856.
- Diene, S.M.; Pinault, L.; Keshri, V.; Armstrong, N.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Colson, P.; La Scola, B.; Rolain, J.-M.; et al. Human metallo-β-lactamase enzymes degrade penicillin. Sci. Rep. 2019, 9, 12173.
- Pettinati, I.; Brem, J.; Lee, S.Y.; McHugh, P.J.; Schofield, C.J. The Chemical Biology of Human Metallo-β-Lactamase Fold Proteins. Trends Biochem. Sci. 2016, 41, 338–355.
- Colson, P.; Pinault, L.; Azza, S.; Armstrong, N.; Chabriere, E.; La Scola, B.; Pontarotti, P.; Raoult, D. A protein of the metallo-hydrolase/oxidoreductase superfamily with both beta-lactamase and ribonuclease activity is linked with translation in giant viruses. Sci. Rep. 2020, 10, 21685.
- Maatouk, M.; Ibrahim, A.; Pinault, L.; Armstrong, N.; Azza, S.; Rolain, J.M.; Bittar, F.; Raoult, D. New Beta-lactamases in Candidate Phyla Radiation: Owning Pleiotropic Enzymes Is a Smart Paradigm for Microorganisms with a Reduced Genome. Int. J. Mol. Sci. 2022, 23, 5446.
- Li, Z.; Huang, H.; Zhao, H.; Meng, K.; Zhao, J.; Shi, P.; Yang, P.; Luo, H.; Wang, Y.; Yao, B. Genetic diversity and expression profiles of cysteine phytases in the sheep rumen during a feeding cycle. Lett. Appl. Microbiol. 2014, 59, 615–620.
- Dersjant-Li, Y.; Awati, A.; Schulze, H.; Partridge, G. Phytase in non-ruminant animal nutrition: A critical review on phytase activities in the gastrointestinal tract and influencing factors. J. Sci. Food Agric. 2015, 95, 878–896.
- Kostelecky, B.; Pohl, E.; Vogel, A.; Schilling, O.; Meyer-Klaucke, W. The crystal structure of the zinc phosphodiesterase from Escherichia coli provides insight into function and cooperativity of tRNase Z-family proteins. J. Bacteriol. 2006, 188, 1607–1614.
- Park, K.S.; Kim, T.Y.; Kim, J.H.; Lee, J.H.; Jeon, J.H.; Karim, A.M.; Malik, S.K.; Lee, S.H. PNGM-1, a novel subclass B3 metallo-β-lactamase from a deep-sea sediment metagenome. J. Glob. Antimicrob. Resist. 2018, 14, 302–305.
- Bergonzi, C.; Schwab, M.; Chabriere, E.; Elias, M. The quorum-quenching lactonase from Alicyclobacter acidoterrestris: Purification, kinetic characterization, crystallization and crystallographic analysis. Acta Cryst. F Struct. Biol. Commun. 2017, 73, 476–480.
- Liu, D.; Lepore, B.W.; Petsko, G.A.; Thomas, P.W.; Stone, E.M.; Fast, W.; Ringe, D. Three-dimensional structure of the quorum-quenching N-acyl homoserine lactone hydrolase from Bacillus thuringiensis. Proc. Natl. Acad. Sci. USA 2005, 102, 11882–11887.
- Mion, S.; Carriot, N.; Lopez, J.; Plener, L.; Ortalo-Magné, A.; Chabrière, E.; Culioli, G.; Daudé, D. Disrupting quorum sensing alters social interactions in Chromobacterium violaceum. npj Biofilms Microbiomes 2021, 7, 40.
- Bergonzi, C.; Schwab, M.; Elias, M. The quorum-quenching lactonase from Geobacillus caldoxylosilyticus: Purification, characterization, crystallization and crystallographic analysis. Acta Crystallogr. Sect. Struct. Biol. Commun. 2016, 72, 681–686.
- Cai, X.; Yu, M.; Shan, H.; Tian, X.; Zheng, Y.; Xue, C.; Zhang, X.H. Characterization of a novel N-acylhomoserine lactonase RmmL from Ruegeria mobilis YJ3. Mar. Drugs 2018, 16, 370.
- Park, S.Y.; Lee, S.J.; Oh, T.K.; Oh, J.W.; Koo, B.T.; Yum, D.Y.; Lee, J.K. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 2003, 149, 1541–1550.
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.H. MomL, a novel marine-derived N-Acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782.
- Wang, W.Z.; Morohoshi, T.; Ikenoya, M.; Someya, N.; Ikeda, T. AiiM, a Novel Class ot N-Acylhomosenne Lactonase from the Leaf-Associated Bacterium Microbacterium testaceum. Appl. Environ. Microbiol. 2010, 76, 2524–2530.
- Wang, W.Z.; Morohoshi, T.; Someya, N.; Ikeda, T. Aidc, a Novel N-Acylhomoserine Lactonase from the Potato Root-Associated Cytophaga-Flavobacteria-Bacteroides (CFB) Group Bacterium Chryseobacterium sp. Strain strb126. Appl. Environ. Microbiol. 2012, 78, 7985–7992.
