Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Controversy continues over the functional prevalence of long non-coding RNAs (lncRNAs) despite their being widely investigated in all kinds of cells and organisms. In animals, lncRNAs have aroused general interest from exponentially increasing transcriptomic repertoires reporting their highly tissue-specific and developmentally dynamic expression, and more importantly, from growing experimental evidence supporting their functionality in facilitating organogenesis and individual fitness.
- lncRNA
- function
- screening
- development
- testis
1. Introduction
Mammalian cells house tens of thousands of long noncoding RNAs (lncRNAs) [1]. By far, dozens of them are well documented as being functional [2][3]. Needless to say, this only represents a minute fraction of the total. Relative to those shown in cell lines, a smaller number of lncRNAs are demonstrated to perform a function required for proper organismal development [4][5]. Even less recognized is the functionality of the mammalian testicular lncRNAs, few of which are fully validated as having a true role in either spermatogenesis or male fertility in knockout mouse models [6], although the testis expresses numerous species of lncRNAs [7][8]. Until now, how large a fraction of lncRNAs represent biological significance remains a central puzzle, fueling a long-lasting debate [9][10][11][12][13][14]. Some researchers argue that even though many useful lncRNAs do exist, the majority are not meaningful, regardless of whether or not they are transcribed excessively [15][16]. This opinion comes from several aspects. From a genomic perspective, recent global transcriptome profiling across eukaryotic organisms demonstrates that at least 85% of the genome is transcribed, which is far more than expected [17]. However, only 10% of the Pol II activities in yeast initiate from conventional promoters and the remaining events are noise [15]. Other evidence comes from several studies using mouse knockout models with the deletion of large DNA fragments that transcribe hundreds of transcripts. Nevertheless, these models reveal no apparent phenotype [18] and many functionless lncRNAs in animal models have been reported [19][20][21]. In contrast, other researchers claim that the functional discovery of lncRNA genes, which are difficult relative to protein-coding genes, is still in its infancy [22]. Still, a repository of functional lncRNAs has been compiled and appreciated [3]. It is thus conceivable that buried in the mass of junk transcripts could be a large proportion of functional lncRNAs that are, nevertheless, emerging like the tip of an iceberg [9][23].
Although much attention has been focused on the discovery of bona fide functional lncRNAs at the organismal level, this field is moving slowly. The human genome harbors a magnitude of lncRNA gene loci, ranging from 59,000 up to 102,000, based on different databases [24][25][26]. About 80% of the human genome is actively transcribed through the body’s development to adulthood [27]. While nearly 2000 lncRNAs have a putative functional association, most of them are described solely in cell lines and have yet to be studied in tissues [3]. To date, only a few lncRNAs have been functionally identified using animal models [2]. A thorough dissection of the functionality of a given lncRNA in organ development calls for effective and multipronged strategies.
2. LncRNA Functionality: Prediction Rationale
With the advent of next-generation RNA sequencing technologies, the expressions of lncRNAs have been subjected to high-throughput analyses in various types of cells or tissues [4][28]. Compared with protein-coding genes, lncRNAs are expressed at lower levels while more specifically, i.e., in a cell type-, tissue-, developmental stage-, or disease state-specific manner [29][30][31]. Most lncRNAs are more tightly regulated than protein-coding genes, contending for a central role of lncRNAs in the cell state determination [32][33]. Thus, discerning functional subsets from a magnitude lncRNA pool is a priority [34] but has become a major hurdle in the field. Screening efforts based on common criteria such as physical gene locus proximity, lncRNA-gene co-expression, and sequence conservation, together with individualized paradigms, have pointed to the role of lncRNA in mammalian organ development [8]. For example, in cis co-expression of lncRNA and nearby genes has been widely applied to predicting functional lncRNAs. However, such a prediction strategy has been challenged in that co-activation of neighboring genes is driven by multiple lncRNA-associated mechanisms, such as enhancer-like DNA locus, splice sites, or transcriptional activity, but not exclusively by the lncRNA transcript itself [35]. Moreover, there are always contrasting variations across different species or organs, leading to generally limited efficacy in picking out bona fide functional lncRNAs [34]. Therefore, reconsidering traditional strategies to achieve a higher probability of mining out functional lncRNAs is now on the horizon.
3. Functional Screening: Cell Lines vs. Animal Models
Benefiting from fast and easy manipulation as well as low cost, cultured cell lines serve as an important platform to decrypt the functionality of individual lncRNAs. Moreover, these advantages make cell lines an ideal system for high-throughput lncRNA functional screening. Indeed, RNAi-, CRISPR/Cas9-, CRISPR/dCas9-, or CRISPR/Cas13-based screening strategies have successfully identified a dozen lncRNAs functioning in cell differentiation, growth, or response to stimuli [36][37][38][39]. While these seminal studies clearly demonstrate the role of lncRNAs, only a small subset of lncRNAs functioning in cell lines could be recapitulated in mouse models. A representative example is Malat1, which regulates growth-control genes at both the transcriptional and post-transcriptional levels [40]. Nevertheless, in vivo knockout mice show normal development and fertility [41]. Another example is Evx1as, which promotes EVX transcription in cis and regulates mesendodermal differentiation in pluripotent cell lines [42]. Nevertheless, Evx1as-ablated mice are viable with no obvious abnormality [19]. Similar contradictory outcomes from cell lines versus animal models are de facto often obtained and discussed in more cases [2].
Regardless of the long manufacturing period and heavy expenditure, the convincing level of lncRNA functional studies requires appropriate LOF manipulation on animals. Indeed, CRISPR/Cas9-based screening using different animal models, including C. elegans, Drosophila, zebrafish, and mouse, has been fruitful, but displays variable results since its emergence. Goudarzi et al. selectively tested 25 candidate lncRNAs based on their conservation, expression trait, and proximity to developmental regulators, showing that lncRNAs have no overt roles in zebrafish [20]. Schor et al. deleted 3 out of 362 lncRNAs with specific spatiotemporal expression in Drosophila embryogenesis but yielded no obvious phenotypes in normal conditions or under stress factors [43]. In addition, the genetic ablation of six cardiac-specific mouse lncRNAs with distinct transcriptional and epigenetic patterns resulted in no defects [44]. Despite these negative screening results in terms of the biological significance of tested lncRNAs, some other screens were more or less successful. Sauvageau et al. filtered out 18 lncRNAs with epigenetic modification for active transcription and conservation between mouse and human, of which three mutant strains displayed lethality and two exhibited growth defects [45]. A large-scale evaluation of 155 C. elegans knockouts of intergenic lncRNAs (lincRNAs) revealed phenotypes of 23 knockouts [46]. Another functional examination of 10 C. elegans knockouts revealed six lncRNAs required for normal development and fertility [47]. As such, two groups of screens led to opinions that could be paradoxical regarding the prevalence of lncRNA functionality in animals, which has rather confused researchers. The in vivo lncRNA functional screens are far less fruitful than the progress in understanding their modes of mechanistic action. This gap is attributed to several explanations [2], including non-specific or off-target effects [48], transcript-independent mechanisms [35][49], functional redundancy [50], non-conserved function among species [51], and a stress- or disease-responsive role [4], as well as a missed phenotype [52][53].
Based on the research discussed above, cell lines and animal models have both advantages and disadvantages in functional screening. A combinational screen using both may allow for attaining a complementarity between large-scale screening ex vivo and the discovery of truly functional lncRNAs in vivo. Perhaps feasibly, for preliminary filtering, high-throughput RNAi or knockout approach can be exploited in primarily derived cell lines and, meanwhile, knockout models can be designed for endogenous validation of potential functional subsets. Indeed, several recent screens applied this dual-means strategy and therefore discovered dozens of lncRNAs in various cell lines and also in their corresponding organs [54][55][56].
4. Testicular LncRNAs: Functionality
Over the past decade, an intense issue has raised the question of how functional lncRNAs originate from pervasive transcription [23][57]. A meaningful explanation is that functional lncRNAs occasionally evolve from a vast pool of non-functional transcripts via a mechanism similar to constructive neutral evolution [23]. In other words, only those organisms that produce a large quantity of junk RNAs possess many functional lncRNAs. Fitting this opinion, lncRNAs are highly expressed in nervous systems, where intensive research has revealed several subsets with physiological roles in neurogenesis, a field moving far beyond others [58].
Actually, the adult testis displays the highest transcriptome complexity among the tissues [8][59][60]. On one hand, during meiosis, chromatin relaxation activates a transcriptional burst of genic and intergenic RNAs, particularly lncRNAs, endo-siRNAs and pachytene piRNAs, along with transpositional shuffling to promote genomic and transcriptomic variability [61]. On the other hand, endo-siRNAs and pachytene piRNAs generated from antisense transcripts, usually from transposon sequences, guide the degradation of mRNAs and lncRNAs that could be useless or even deleterious, referred to as a molecular mechanism for genome-wide quality control [61][62]. Combined, the extensive transcription and targeted degradation of lncRNAs accelerate lncRNA evolution under robust screening through natural selection filters in mammalian spermatogenesis, giving rise to the thriving birth of young lncRNAs and preservation of optimized ones thereof [7][61]. All these clues highlight the significance of lncRNAs in spermatogenesis and hint at the existence of not a few functional ones. Although massive novel spermatogenic lncRNAs have been identified, most of them are biologically uncharacterized.
Several studies show the role of lncRNAs in male germ cells, which was summarized in recent reviews [6][63][64][65]. So far, researchers still know little about lncRNA function in spermatogenesis, especially in mammals. There are only four lncRNA knockout models displaying obvious phenotypes (Figure 1). The first case is Tsx (testis-specific X-linked), whose knockout males were fertile with normal spermatogenesis [66]. Nonetheless, Tsx knockout resulted in a mild increase in the apoptosis of pachytene spermatocytes, an additional maternal-specific effect on litter size, and dysfunction of multiple other cell types. The second case is Tesh1, whose knockout males were subfertile with teratospermia and offspring with female-biased sex ratios [67]. The third case is Tug1 (taurine-up-regulated gene 1), whose knockout males were sterile with decreased sperm counts and malformed sperm morphologies [68]. However, Tug1 may not act merely as a lncRNA transcript because there are two additional attributive layers: as a cis-DNA regulator and as a protein-coding gene [68]. Researchers still know little about whether these mechanisms alone or jointly contribute to its function in spermatogenesis [22]. The last case is Gm9999, which encodes two small peptides named Kastor and Polluks, respectively. Deletion of both peptides caused male subfertility with teratospermia and deletion of one peptide partially reproduced the phenotype, suggesting that Gm9999 regulates spermatogenesis on the dependence of peptide generation [69]. These four rarely reported knockout mouse models that exhibit either mild or non-lncRNA transcript-exclusive effects on male fertility implicate a general difficulty in decoding the functional tacitness and complexity of testicular lncRNAs.
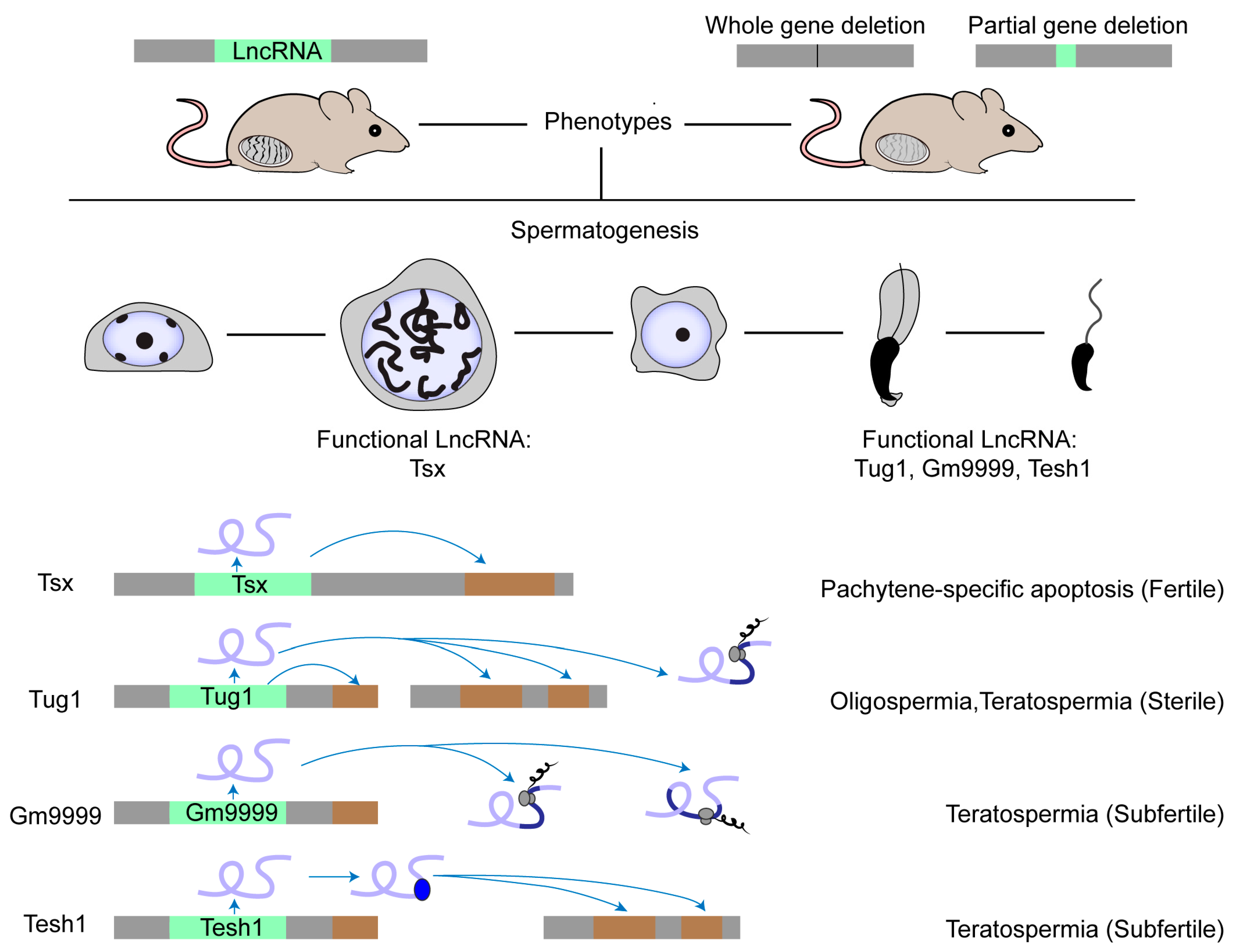
Figure 1. Physiologically functional lncRNAs in mouse testis. The physiological role of a few lncRNAs in mouse spermatogenesis has been studied by creating knockout mouse models. Most lncRNA knockouts delete the entire genomic locus. Other knockouts delete the functional element in the lncRNA locus, such as the ORF region in the Gm9999 locus. So far, only four lncRNAs have been demonstrated as functional in mouse testis. Tsx is highly expressed in pachytene spermatocytes and Tesh1 is mainly expressed in elongated spermatids. Tug1, Gm9999, and Tesh1 knockouts exhibit teratospermia and impaired male fertility. Mechanistically, Tsx acts in cis and Tesh1 in trans. Tug1 could act in cis, in trans, or by an encoded protein. Gm9999 executes its function through its two encoded polypeptides.
5. Testicular LncRNAs: Functional Screening
In one systematic study consisting of the functional screening of testicular lncRNAs, Wen et al. identified 128 testis-specific Drosophila lncRNAs and knocked out 105 of them [70]. Among these knockouts, 33 (31%) showed visual developmental defects in late male germ cells and a partial or complete loss of male fertility. This result supports the pervasive involvement of functional lncRNAs, at least in late spermatogenesis, and for the first time, supports a general relevance of lncRNAs to testis function. This study employed RNAi and rescue experiments to determine in cis or in trans regulation of gene expression and also to separate RNA-dependent lncRNA function from DNA-dependent effect. From an evolutionary viewpoint, a constant proportion (around 30% of the entire pool) of functional lncRNA sequences are likely accumulating all along, nearly equal in percentage to this study [70]. In another functional screen in mouse testis, Li et al. conducted global lncRNA expression profiling in six types of spermatogenic cells and subsequent filtering was based on protein-coding gene co-expression analysis and physical proximity with spermatogenic regulators [71]. They finally selected six lncRNA candidates and employed an shRNA-knockdown system in the testis to interrogate their in vivo function. As a result, two out of six candidate-depleted testes exhibited overt developmental defects in spermatogenesis. Fascinatingly, these screens in the testis of different species seem more efficient than in other tissues, supporting the aforementioned assumption that testicular transcriptome may comprise a relatively larger pool of functional lncRNAs.
This entry is adapted from the peer-reviewed paper 10.3390/ncrna9040036
References
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigo, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548.
- Gao, F.; Cai, Y.; Kapranov, P.; Xu, D. Reverse-genetics studies of lncRNAs-what we have learnt and paths forward. Genome Biol. 2020, 21, 93.
- Ma, L.; Cao, J.; Liu, L.; Du, Q.; Li, Z.; Zou, D.; Bajic, V.B.; Zhang, Z. LncBook: A curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019, 47, 2699.
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21.
- Rinn, J.L.; Chang, H.Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 2020, 89, 283–308.
- Joshi, M.; Rajender, S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod. Biol. Endocrinol. RBE 2020, 18, 103.
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grutzner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640.
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019, 571, 510–514.
- Jandura, A.; Krause, H.M. The New RNA World: Growing Evidence for Long Noncoding RNA Functionality. Trends Genet. TIG 2017, 33, 665–676.
- Brosius, J. Waste not, want not-transcript excess in multicellular eukaryotes. Trends Genet. TIG 2005, 21, 287–288.
- Semon, M.; Duret, L. Evidence that functional transcription units cover at least half of the human genome. Trends Genet. TIG 2004, 20, 229–232.
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2.
- Doolittle, W.F. We simply cannot go on being so vague about ‘function’. Genome Biol. 2018, 19, 223.
- Kowalczyk, M.S.; Higgs, D.R.; Gingeras, T.R. Molecular biology: RNA discrimination. Nature 2012, 482, 310–311.
- Struhl, K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007, 14, 103–105.
- Palazzo, A.F.; Gregory, T.R. The case for junk DNA. PLoS Genet. 2014, 10, e1004351.
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013, 9, e1003569.
- Nobrega, M.A.; Zhu, Y.; Plajzer-Frick, I.; Afzal, V.; Rubin, E.M. Megabase deletions of gene deserts result in viable mice. Nature 2004, 431, 988–993.
- Han, X.; Luo, S.; Peng, G.; Lu, J.Y.; Cui, G.; Liu, L.; Yan, P.; Yin, Y.; Liu, W.; Wang, R.; et al. Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J. Mol. Cell Biol. 2018, 10, 175–178.
- Goudarzi, M.; Berg, K.; Pieper, L.M.; Schier, A.F. Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility. eLife 2019, 8, e40815.
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087.
- Andergassen, D.; Rinn, J.L. From genotype to phenotype: Genetics of mammalian long non-coding RNAs in vivo. Nat. Rev. Genet. 2022, 23, 229–243.
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161.
- Frankish, A.; Diekhans, M.; Ferreira, A.M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019, 47, D766–D773.
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208.
- Zhao, Y.; Li, H.; Fang, S.; Kang, Y.; Wu, W.; Hao, Y.; Li, Z.; Bu, D.; Sun, N.; Zhang, M.Q.; et al. NONCODE 2016: An informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016, 44, D203–D208.
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Deveson, I.W.; Hardwick, S.A.; Mercer, T.R.; Mattick, J.S. The Dimensions, Dynamics, and Relevance of the Mammalian Noncoding Transcriptome. Trends Genet. TIG 2017, 33, 464–478.
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes. Dev. 2011, 25, 1915–1927.
- Pauli, A.; Valen, E.; Lin, M.F.; Garber, M.; Vastenhouw, N.L.; Levin, J.Z.; Fan, L.; Sandelin, A.; Rinn, J.L.; Regev, A.; et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 2012, 22, 577–591.
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346.
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227.
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62.
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407.
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455.
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300.
- Stojic, L.; Lun, A.T.L.; Mangei, J.; Mascalchi, P.; Quarantotti, V.; Barr, A.R.; Bakal, C.; Marioni, J.C.; Gergely, F.; Odom, D.T. Specificity of RNAi, LNA and CRISPRi as loss-of-function methods in transcriptional analysis. Nucleic Acids Res. 2018, 46, 5950–5966.
- Bester, A.C.; Lee, J.D.; Chavez, A.; Lee, Y.R.; Nachmani, D.; Vora, S.; Victor, J.; Sauvageau, M.; Monteleone, E.; Rinn, J.L.; et al. An Integrated Genome-wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell 2018, 173, 649–664.e620.
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355, eaah7111.
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Non-Coding RNA 2020, 6, 22.
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012, 2, 111–123.
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652.
- Schor, I.E.; Bussotti, G.; Males, M.; Forneris, M.; Viales, R.R.; Enright, A.J.; Furlong, E.E.M. Non-coding RNA Expression, Function, and Variation during Drosophila Embryogenesis. Curr. Biol. CB 2018, 28, 3547–3561.e3549.
- George, M.R.; Duan, Q.; Nagle, A.; Kathiriya, I.S.; Huang, Y.; Rao, K.; Haldar, S.M.; Bruneau, B.G. Minimal in vivo requirements for developmentally regulated cardiac long intergenic non-coding RNAs. Development 2019, 146, dev185314.
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2013, 2, e01749.
- Wei, S.; Chen, H.; Dzakah, E.E.; Yu, B.; Wang, X.; Fu, T.; Li, J.; Liu, L.; Fang, S.; Liu, W.; et al. Systematic evaluation of C. elegans lincRNAs with CRISPR knockout mutants. Genome Biol. 2019, 20, 7.
- Akay, A.; Jordan, D.; Navarro, I.C.; Wrzesinski, T.; Ponting, C.P.; Miska, E.A.; Haerty, W. Identification of functional long non-coding RNAs in C. elegans. BMC Biol. 2019, 17, 14.
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683.
- Paralkar, V.R.; Taborda, C.C.; Huang, P.; Yao, Y.; Kossenkov, A.V.; Prasad, R.; Luan, J.; Davies, J.O.; Hughes, J.R.; Hardison, R.C.; et al. Unlinking an lncRNA from Its Associated cis Element. Mol. Cell 2016, 62, 104–110.
- Meller, V.H.; Rattner, B.P. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002, 21, 1084–1091.
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636.e622.
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39.
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627.
- Perry, R.B.; Hezroni, H.; Goldrich, M.J.; Ulitsky, I. Regulation of Neuroregeneration by Long Noncoding RNAs. Mol. Cell 2018, 72, 553–567.e555.
- Ponnusamy, M.; Liu, F.; Zhang, Y.H.; Li, R.B.; Zhai, M.; Liu, F.; Zhou, L.Y.; Liu, C.Y.; Yan, K.W.; Dong, Y.H.; et al. Long Noncoding RNA CPR (Cardiomyocyte Proliferation Regulator) Regulates Cardiomyocyte Proliferation and Cardiac Repair. Circulation 2019, 139, 2668–2684.
- Zhu, P.; Wu, J.; Wang, Y.; Zhu, X.; Lu, T.; Liu, B.; He, L.; Ye, B.; Wang, S.; Meng, S.; et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat. Cell Biol. 2018, 20, 1134–1144.
- Koonin, E.V. The meaning of biological information. Philos. Trans. Ser. A Math. Phys. Eng. Sci. 2016, 374, 20150065.
- Briggs, J.A.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of Long Non-coding RNAs in Mammalian Nervous System Development, Plasticity, Disease, and Evolution. Neuron 2015, 88, 861–877.
- Necsulea, A.; Kaessmann, H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 2014, 15, 734–748.
- Soumillon, M.; Necsulea, A.; Weier, M.; Brawand, D.; Zhang, X.; Gu, H.; Barthes, P.; Kokkinaki, M.; Nef, S.; Gnirke, A.; et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013, 3, 2179–2190.
- Werner, A.; Piatek, M.J.; Mattick, J.S. Transpositional shuffling and quality control in male germ cells to enhance evolution of complex organisms. Ann. N. Y. Acad. Sci. 2015, 1341, 156–163.
- Watanabe, T.; Cheng, E.C.; Zhong, M.; Lin, H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015, 25, 368–380.
- Zhang, C.; Gao, L.; Xu, E.Y. LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development. Semin. Cell Dev. Biol. 2016, 59, 110–117.
- Luk, A.C.; Chan, W.Y.; Rennert, O.M.; Lee, T.L. Long noncoding RNAs in spermatogenesis: Insights from recent high-throughput transcriptome studies. Reproduction 2014, 147, R131–R141.
- Tzur, Y.B. lncRNAs in fertility: Redefining the gene expression paradigm? Trends Genet. TIG 2022, 38, 1170–1179.
- Anguera, M.C.; Ma, W.; Clift, D.; Namekawa, S.; Kelleher, R.J., III; Lee, J.T. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011, 7, e1002248.
- Hong, S.H.; Han, G.; Lee, S.J.; Cocquet, J.; Cho, C. Testicular germ cell-specific lncRNA, Teshl, is required for complete expression of Y chromosome genes and a normal offspring sex ratio. Sci. Adv. 2021, 7, eabg5177.
- Lewandowski, J.P.; Dumbovic, G.; Watson, A.R.; Hwang, T.; Jacobs-Palmer, E.; Chang, N.; Much, C.; Turner, K.M.; Kirby, C.; Rubinstein, N.D.; et al. The Tug1 lncRNA locus is essential for male fertility. Genome Biol. 2020, 21, 237.
- Mise, S.; Matsumoto, A.; Shimada, K.; Hosaka, T.; Takahashi, M.; Ichihara, K.; Shimizu, H.; Shiraishi, C.; Saito, D.; Suyama, M.; et al. Kastor and Polluks polypeptides encoded by a single gene locus cooperatively regulate VDAC and spermatogenesis. Nat. Commun. 2022, 13, 1071.
- Wen, K.; Yang, L.; Xiong, T.; Di, C.; Ma, D.; Wu, M.; Xue, Z.; Zhang, X.; Long, L.; Zhang, W.; et al. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res. 2016, 26, 1233–1244.
- Li, K.; Xu, J.; Luo, Y.; Zou, D.; Han, R.; Zhong, S.; Zhao, Q.; Mang, X.; Li, M.; Si, Y.; et al. Panoramic transcriptome analysis and functional screening of long noncoding RNAs in mouse spermatogenesis. Genome Res. 2021, 31, 13–26.
This entry is offline, you can click here to edit this entry!
