Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
One of the strategies to overcome diseases or abiotic stress in crops is the use of improved varieties. Genetic improvement could be accomplished through different methods, including conventional breeding, induced mutation, genetic transformation, or gene editing. The gene function and regulated expression through promoters are necessary for transgenic crops to improve specific traits. The variety of promoter sequences has increased in the generation of genetically modified crops because they could lead to the expression of the gene responsible for the improved trait in a specific manner.
- transcription
- gene expression
- genetic engineering
1. Promoters and Their Importance in Genetically Modified Crops
Sustainable maintenance in agriculture with a high productivity level has also been thanks to advances in plant biotechnology, especially through the implementation of genetically modified crops. Therefore, the search and availability of new promoters have been a high priority among researchers since they provide spatial and temporal control in transgene expression. Over the years, various promoters have been identified in different organisms for use in genetically modified plants because they are a potent tool in regulating gene expression leading to improved traits [1]. The success of gene expression depends to a great extent on the use and efficient selection of promoters, even being able to drive multiple transgenes by the same promoter in a single plant [2][3].
Promoters are located in the 5′ region of a gene and are composed of a specific nucleotide sequence controlling the expression of DNA in a physical, adjacent, and functional manner. In this way, gene regulation mainly depends on these essential regulatory elements for transcription initiation [1]. A DNA sequence located at the 5′ end of the coding region of a gene, which includes different binding regions for transcription factors, is known as a promoter [4]. Promoters are divided into a central core and several regulatory regions, usually in the 5′ region. The promoter core may also have a TATA box (consensus DNA sequence rich in adenine and thymine) and an initiator element that binds to transcription factors. It is already known that promoters can regulate the expression level of various transgenes and, in turn, these can be obtained from different sources; thus, their classification is divided into pol II (constitutive, inducible, and tissue-specific) and pol III (U3, U6); both are activated due to recognition by RNA polymerases [5].
On the other hand, the initiator elements can signal the start of the transcription in specific promoters that lack a TATA box [6]. The recognition of plant promoters would generally involve identifying and characterizing genes expressed in tissues or under conditions of some physiological stress [7] to identify promoters activated in these circumstances. The structurally characterized promoters could be fused with a coding sequence for later use in the genetic transformation of plants [8]; thus, the promoters are necessary to determine the expression of transgenes in genetically modified crops (Figure 1).
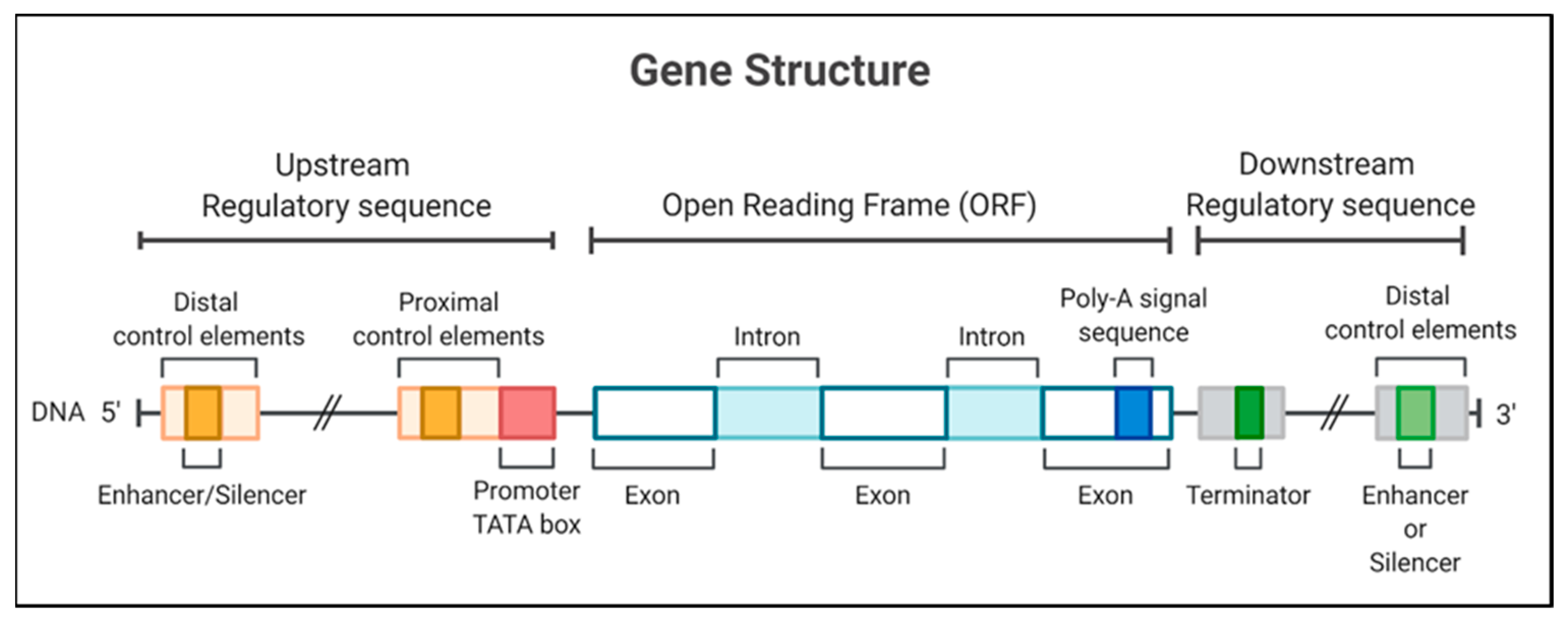
Figure 1. Gene structure, including the promoter region for a eukaryotic gene, encodes a protein. The gene diagrammed here contains a TATA box and different regulatory elements in 5′ and 3′ regions.
The study of promoters is necessary to understand the regulation of gene expression in plants [9]. The isolation of genes could be used as a starting point for identifying the promoters which drive the expression of the isolated gene. If the genome of the organism is already sequenced, basic local alignment tool (blast) analysis could be performed using the query of the sequence of the isolated gene; then, the location of the promoters will be at the 5′ region of the coding sequence of the gene. In the case that the genome of an organism is not sequenced, first, genes could be isolated through the creation of libraries, and once the gene sequence is known, the promoters of corresponding genes could be searched using genome walking strategies, such as PCR-based techniques [10][11]. The importance of isolating and identifying promoters lies in knowing which regions of interest are located on the 5′ side of a known genomic sequence.
Whole-genome sequencing of many plants has facilitated the isolation of promoters from genomes, in which primers can be designed in the 5′ region of the open reading frame [12]. Once the promoter is isolated and sequenced, in silico analysis can be performed using bioinformatics tools such as PlantCARE [13], PlantProm DB [14], iProm-Zea [15], and TSSPlant [16]. Recent studies have shown that promoters play an essential role in processes such as gene editing with clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9), increasing the effectiveness and specificity in cutting double-stranded DNA. For example, in soybean, the function of the gRNA and Cas9 expressed under the control of the double 35S Cauliflower Mosaic Virus (CaMV) promoter was tested in hairy soybean roots where a high rate of mutations was obtained; which were even visualized in the next generation; opposite when using egg cell-specific promoters [17][18].
2. Types of Promoters
Promoters can be classified as constitutive, tissue-specific, inducible, and synthetic. The classification will depend on the function, type, and gene expression level [13][14].
3. Constitutive Promoters
Constitutive promoters are primarily used in plant transformation, promoting the expression of transgenes with different purposes, whether resistant to diseases, biotic stress, or insects [19]. One of the characteristics of the constitutive promoters is that they maintain control of the genes during most of the plant development; the expression levels will depend clearly on the type of cell with which they work. The 35S promoter derived from the CaMV is genetically modified crops’ most popular constitutive promoter (Table 1).
Table 1. Constitutive promoters are used in the generation of genetically modified plants.
| Promoter | Source | Expression | References |
|---|---|---|---|
| Act1 | Actin gene, rice | The whole plant, preferably monocots | [20][21] |
| Adh1 | Alcohol dehydrogenase gene, maize | Roots, meristematic tissue, endosperm, and pollen (anaerobic regulation) preference in monocots | [22][23] |
| HSP18.2 | Arabidopsis thaliana | Leaves, vascular system | [24] |
| ScBV | Bacilliform virus, sugarcane | Leaves, vascular system, monocots, and dicots | [25][26] |
| Ubi-1 | Ubiquitin gene, maize | Protoplast, monocots. | [27] |
| RUBQ1/RUBQ2 | Ubiquitin gene, rice | Genes expression, monocots. | [28] |
| Gmubi | Soybean | The whole plant | [29] |
| CaMV35S | Cauliflower mosaic virus | Expression throughout the plant, monocots, and dicots | [30] |
| nos | Nopaline synthase gene, Agrobacterium tumefaciens | Expression throughout the plant, monocots, and dicots | [31][32] |
| CmYLCV | Cestrum Yellow Leaf Curling Virus (CmYLCV) | Growth and development | [33] |
| KST1 | Solanum tuberosum (potato) | Guard cell promoter | [34] |
| Cula11/Cula08 | Cunninghamia lanceolate (Chinese fir) | Protoplast, monocots, and dicots | [35] |
| P OsCon1 | Rice | The whole plant, monocots, and dicots | [36] |
| TCTP | Oil palm | Immature embryo, embryogenic callus, embryoid, a young leaflet from a mature palm, green leaf, mesocarp, and stem | [37] |
The function of the CaMV35S promoter was analyzed in cell colonies and in vitro banana plants by fusion with reporter genes (luciferase and β-glucuronidase) and the subsequent transformation of banana embryogenic cell suspensions (ECS) [38]. This promoter has been fused to the luc2 reporter gene (isolated from the vector pGL4.10, Promega). Its activity has been characterized in cell lines and in vitro genetically modified banana plants [39][40]. The native banana promoter demonstrates high activity in ‘Williams’ banana plants in vitro (Figure 2).
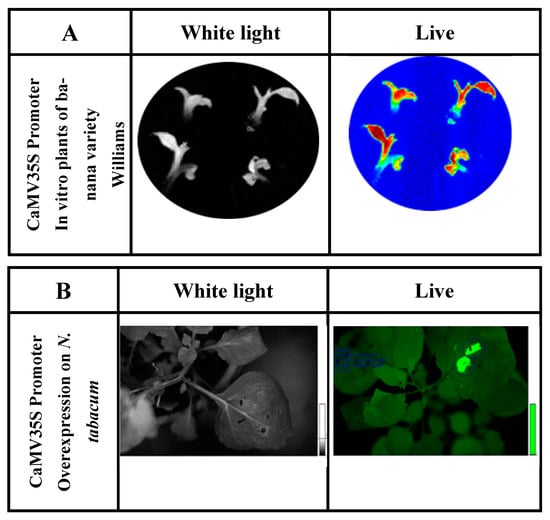
Figure 2. (A) Determination of luciferase reporter gene activity of in vitro ‘Williams’ banana plants transformed with the construct containing P35S::luc2. Exposure time was 3 min in total darkness; the photo was captured using the Stella 3200 equipment camera (Raytest, Straubenhardt, Germany). Luciferin dissolved in water (500 µm) was applied to in vitro plants. ‘Live’ indicates luciferase enzyme activity detected under conditions of total darkness. (B) Luciferase activity in N. tabacum leaves agroinfiltrated for the luciferase reporter gene expression fused to the CaMV 35S promoter (P35S::luc2).
On the other hand, the characterization of the constitutive actin promoter demonstrated its efficacy in regulating genes in rice protoplasts when fused to the β-glucuronidase (gus) gene [20]. Like the study of ubiquitin genes, it has provided highly expressed plant constitutive promoters, especially in monocotyledonous plants isolated from rice [41] and maize [42].
4. Tissue-Specific Promoters
In the case of specific promoters, these direct the gene expression in a particular tissue, which can happen in different parts of the plant and at different stages [43]. In plant tissue, promoters can control/initiate the expression of a specific gene, such as in roots, seeds, or the vascular system [6].
For the expression of specific genes in genetically modified crops, it has been decided to use native or homologous promoters since they present a high specificity guaranteeing a correct genetic transformation [44]. In this concept, Manavella and Chan [45] mention the importance of using tissue-specific promoters since it would allow for more efficient crops [45] because the programmed regulation of the promoters is due to the correct expression of the transcription factors [43]. In this way, the study of vitamin A has been a public health problem worldwide, which is why Paul et al. [46] performed the characterization and isolation of two banana promoters of the expansin1 gene (Exp1) and a fruit-specific oxidase (ACO) promoter for increasing vitamin A or retinol. These promoters were fused with the gus gene, and the activity of the fruit pulp was quantified by an ELISA assay, while fluorometric assays were used for the leaves and peel to determine the enzymatic activity. Finally, it was possible to determine that the prolonged maturation time of this fruit is essential to obtaining optimal concentrations of vitamin A. Few tissue-specific promoters have tangible expressions; for instance, Ye et al. [47] characterized two new cis-regulatory elements, GSE1 and GSE2. GSE1 activates promoters in all green plant tissues, while GSE2 is a regulator in the sheath and stem of plants, weakening gene expression [47].
Trivedi and Nath [48] performed the characterization and identification of the Expansin promoter for the first time, concluding that exposure to ethylene induces the overexpression of MaEXPA1, which progressively increases in the fruit and does not present expression in any other plant tissue. Furthermore, another four banana promoters were analyzed during maturation exposed to ethylene (MaEXPA2, MaEXPA3, MaEXPA4, and MaEXPA5) for the study in both dicots and monocots [48].
Only the MaEXPA2 promoter was specific for the fruit and had a more robust expression, which is why it is recommended for application in monocots. In contrast, the other promoters were expressed in different plant tissues, although their competence could be analyzed in dicotyledonous plants [49]. The MaEXPA1 promoter has already been characterized and is active in fruit tissue.
This entry is adapted from the peer-reviewed paper 10.3390/genes14061226
References
- Potenza, C.; Aleman, L.; Sengupta-Gopalan, C. Targeting transgene expression in research, agricultural, and en-vironmental applications: Promoters used in plant transformation. Vitr. Cell. Dev. Biol. Plant 2004, 40, 1–22.
- Danila, F.; Schreiber, T.; Ermakova, M.; Hua, L.; Vlad, D.; Lo, S.; Chen, Y.; Lambret-Frotte, J.; Hermanns, A.S.; Athmer, B.; et al. A single promoter-TALE system for tissue-specific and tuneable expression of multiple genes in rice. Plant Biotechnol. J. 2022, 20, 1786–1806.
- Dafny-Yelin, M.; Tzfira, T. Delivery of Multiple Transgenes to Plant Cells. Plant Physiol. 2007, 145, 1118–1128.
- Southern, M.M.; Brown, P.E.; Hall, A. Luciferases as Reporter Genes. Methods Mol. Biol. 2006, 323, 293–306.
- Kor, S.D.; Chowdhury, N.; Keot, A.K.; Yogendra, K.; Chikkaputtaiah, C.; Reddy, P.S. RNA Pol III promoters—Key players in precisely targeted plant genome editing. Front. Genet. 2023, 13, 989199.
- Dutt, M.; Dhekney, S.A.; Soriano, L.; Kandel, R.; Grosser, J.W. Temporal and spatial control of gene expression in horticultural crops. Hortic. Res. 2014, 1, 14047.
- Cai, M.; Wei, J.; Li, X.; Xu, C.; Wang, S. A rice promoter containing both novel positive and negative cis-elements for regulation of green tissue-specific gene expression in transgenic plants. Plant Biotechnol. J. 2007, 5, 664–674.
- Porto, M.S.; Pinheiro, M.P.N.; Batista, V.G.L.; Dos Santos, R.C.; De Albuquerque Melo Filho, P.; De Lima, L.M. Plant promoters: An approach of structure and function. Mol. Biotechnol. 2014, 56, 38–49.
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119.
- Damaj, M.B.; Beremand, P.D.; Buenrostro-Nava, M.T.; Ivy, J.; Kumpatla, S.P.; Jifon, J.; Beyene, G.; Yu, Q.; Thomas, T.L.; Mirkov, T.E. Isolating promoters of multigene family members from the polyploid sugarcane genome by PCR-based walking in BAC DNA. Genome 2010, 53, 840–847.
- Devic, M.; Albert, S.; Delseny, M.; Roscoe, T.J. Efficient PCR walking on plant genomic DNA. Plant Physiol. Biochem. 1997, 35, 331–339.
- Rishi, A.S.; Nelson, N.D.; Goyal, A. Genome walking of large fragments: An improved method. J. Biotechnol. 2004, 111, 9–15.
- Santos, E.; Pacheco, R.; Villao, L.; Galarza, L.; Ochoa, D.; Jordán, C.; Flores, J. Promoter analysis in banana. In Banana: Genomics and Transgenic Approaches for Genetic Improvement; Springer: Singapore, 2016; pp. 157–179.
- Shahmuradov, I.A.; Gammerman, A.J.; Hancock, J.M.; Bramley, P.M.; Solovyev, V.V. PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 2003, 31, 114–117.
- Kim, J.; Shujaat, M.; Tayara, H. iProm-Zea: A two-layer model to identify plant promoters and their types using convolutional neural network. Genomics 2022, 114, 110384.
- Shahmuradov, I.A.; Umarov, R.; Solovyev, V.V. TSSPlant: A new tool for prediction of plant Pol II promoters. Nucleic Acids Res. 2017, 45, e65.
- Zheng, N.; Li, T.; Dittman, J.D.; Su, J.; Li, R.; Gassmann, W.; Peng, D.; Whitham, S.A.; Liu, S.; Yang, B. CRISPR/Cas9-Based Gene Editing Using Egg Cell-Specific Promoters in Arabidopsis and Soybean. Front. Plant Sci. 2020, 11, 800.
- Jiang, F.; Doudna, J.A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529.
- Chen, Z.; Wang, J.; Ye, M.X.; Li, H.; Ji, L.X.; Li, Y.; Cui, D.Q.; Liu, J.M.; An, X.M. A Novel Moderate Constitutive Promoter Derived from Poplar (Populus tomentosa Carrière). Int. J. Mol. Sci. 2013, 14, 6187.
- McElroy, D.; Zhang, W.; Cao, J.; Wu, R. Isolation of an Efficient Actin Promoter for Use in Rice Transformation. Plant Cell 1990, 2, 163.
- Wang, Y.; Zhang, W.; Cao, J.; McElroy, D.; Wu, R. Characterization of cis-acting elements regulating transcription from the promoter of a constitutively active rice actin gene. Mol. Cell Biol. 1992, 12, 3399–3406.
- Kyozuka, J.; Fujimoto, H.; Izawa, T.; Shimamoto, K. Anaerobic induction and tissue-specific expression of maize Adh1 promoter in transgenic rice plants and their progeny. Mol. Genet. Genom. 1991, 228, 40–48.
- Onnela, M.-L.; Suihko, M.-L.; Penttilä, M.; Keränen, S. Use of a modified alcohol dehydrogenase, ADH1, promoter in construction of diacetyl non-producing brewer’s yeast. J. Biotechnol. 1996, 49, 101–109.
- Takahashi, T.; Naito, S.; Komeda, Y. The Arabidopsis HSP18.2 promoter/GUS gene fusion in transgenic Arabidopsis plants: A powerful tool for the isolation of regulatory mutants of the heat-shock response. Plant J. 1992, 2, 751–761.
- Tzafrir, I.; Torbert, K.A.; Lockhart, B.E.; Somers, D.A.; Olszewski, N.E. The sugarcane bacilliform badnavirus promoter is active in both monocots and dicots. Plant Mol. Biol. 1998, 38, 347–356.
- Schenk, P.M.; Sagi, L.; Remans, T.; Dietzgen, R.G.; Bernard, M.J.; Graham, M.W.; Manners, J.M. A promoter from sugarcane bacilliform badnavirus drives transgene expression in banana and other monocot and dicot plants. Plant Mol. Biol. 1999, 39, 1221–1230.
- Christensen, A.H.; Sharrock, R.A.; Quail, P.H. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 1992, 18, 675–689.
- Wang, J.; Oard, J.H. Rice ubiquitin promoters: Deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 2003, 22, 129–134.
- Hernandez-Garcia, C.M.; Martinelli, A.P.; Bouchard, R.A.; Finer, J.J. A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep. 2009, 28, 837–849.
- Odell, J.T.; Nagy, F.; Chua, N.-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812.
- An, G.; Costa, M.A.; Ha, S.B. Nopaline synthase promoter is wound inducible and auxin inducible. Plant Cell 1990, 2, 225–233.
- Bevan, M.; Barnes, W.M.; Chilton, M.-D. Strcuture and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 1983, 11, 369–385.
- Sahoo, D.K.; Sarkar, S.; Maiti, I.B.; Dey, N. Novel Synthetic Promoters from the Cestrum Yellow Leaf Curling Virus. Methods Mol. Biol. 2016, 1482, 111–138.
- Kelly, G.; Lugassi, N.; Belausov, E.; Wolf, D.; Khamaisi, B.; Brandsma, D.; Kottapalli, J.; Fidel, L.; Ben-Zvi, B.; Egbaria, A.; et al. The Solanum tuberosum KST1 partial promoter as a tool for guard cell expression in multiple plant species. J. Exp. Bot. 2017, 68, 2885–2897.
- Ye, S.; Ding, W.; Bai, W.; Lu, J.; Zhou, L.; Ma, X.; Zhu, Q. Application of a novel strong promoter from Chinese fir (Cunninghamia lanceolate) in the CRISPR/Cas mediated genome editing of its protoplasts and transgenesis of rice and poplar. Front Plant Sci. 2023, 14, 1289.
- Li, J.; Xu, R.F.; Qin, R.Y.; Ma, H.; Li, H.; Zhang, Y.P.; Li, L.; Wei, P.C.; Yang, J.B. Isolation and functional characterization of a novel rice constitutive promoter. Plant Cell Rep. 2014, 33, 1651–1660.
- Masura, S.S.; Parveez, G.K.A.; Ti, L.L.E. Isolation and characterization of an oil palm constitutive promoter derived from a translationally control tumor protein (TCTP) gene. Plant Physiol. Biochem. 2011, 49, 701–708.
- Santos, E.; Remy, S.; Thiry, E.; Windelinckx, S.; Swennen, R.; Sági, L. Characterization and isolation of a T-DNA tagged banana promoter active during in vitro culture and low temperature stress. BMC Plant Biol. 2009, 9, 100187.
- Villao, L.; Sánchez, E.; Romero, C.; Galarza, L.; Flores, J.; Santos-Ordóñez, E. Activity characterization of the plantain promoter from the heavy metal-associated isoprenylated plant gene (MabHIPP) using the luciferase reporter gene. Plant Gene 2019, 19, 100187.
- Villao, L.; Flores, J.; Santos-Ordóñez, E. Genetic transformation of apical meristematic shoots in the banana cultivar ‘Williams’. Bionatura 2021, 6, 1462–1465.
- Park, S.H.; Yi, N.; Kim, Y.S.; Jeong, M.H.; Bang, S.W.; Choi YDo Kim, J.K. Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 2010, 61, 2459.
- Cornejo, M.J.; Luth, D.; Blankenship, K.M.; Anderson, O.D.; Blechl, A.E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 1993, 23, 567–581.
- Marín-Rodríguez, M.C.; Smith, D.L.; Manning, K.; Orchard, J.; Seymour, G.B. Pectate lyase gene expression and enzyme activity in ripening banana fruit. Plant Mol. Biol. 2003, 51, 851–857.
- Nazir, S.; Iqbal, M.Z.; Rahman, S.U. Molecular Identification of Genetically Modified Crops for Biosafety and Legitimacy of Transgenes. In Gene Editing-Technologies and Applications; IntechOpen: London, UK, 2019.
- Manavella, P.A.; Chan, R.L. Development of Tissue-Specific Promoters for Plant Transformation. Plant Physiol. 2006, 2, 21–25.
- Paul, J.Y.; Khanna, H.; Kleidon, J.; Hoang, P.; Geijskes, J.; Daniells, J.; Zaplin, E.; Rosenberg, Y.; James, A.; Mlalazi, B.; et al. Golden bananas in the field: Elevated fruit pro-vitamin A from the expression of a single banana transgene. Plant Biotechnol. J. 2017, 15, 520–532.
- Ye, R.; Zhou, F.; Lin, Y. Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L. ssp.). Plant Cell Rep. 2012, 31, 1159–1172.
- Trivedi, P.K.; Nath, P. MaExp1, an ethylene-induced expansin from ripening banana fruit. Plant Sci. 2004, 167, 1351–1358.
- Sane, V.A.; Sane, A.P.; Nath, P. Multiple forms of α-expansin genes are expressed during banana fruit ripening and development. Postharvest Biol. Technol. 2007, 45, 184–192.
This entry is offline, you can click here to edit this entry!
