Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
There is a significant body of evidence that has demonstrated that Signal Transducers and Activators of Transcription (STATs) play a critical role in ovarian cancer progression. The STAT family is comprised of seven distinct proteins, STAT1, STAT2, STAT3, STAT4, STAT5A and 5B, and STAT6. The structure among STAT proteins remains highly conserved, with all STAT proteins encoding a coiled-coil (CC) domain, a DNA-binding (DB) domain, and a SRC homology 2 (SH2) domain.
- STAT
- signaling
- isoforms
- immune cell
- targeted therapy
1. Structure of STATs
STAT proteins are highly conserved, with the molecular weights of full-length STAT proteins being around 80–100 kDa [1]. As described in Figure 1, at the N-terminus, STATs contain an N-terminal domain (NTD), which is followed by a coiled-coil (CC) domain. The DNA-binding (DB) domain is located near the center of the protein and is separated from the SH2 domain by a linker (LD). At the c-terminus of the protein, there is a trans-activating domain (TAD) [2]. The NTD has importance in STAT protein interactions, including STAT dimers, transcription factors, cofactors, and receptors [3]. The CC domain, which encodes four-helix bundles is involved in protein–protein interactions that control nuclear import and export processes [3]. Moreover, while the DB domain is important for binding the chromatin and inducing transcription, it also encodes sequences that are essential for nuclear import. Moreover, in addition to providing structural organization, the linker domain plays a role in transcriptional regulation of STAT1 [4]. The SH2 domain primarily functions in recognizing phosphotyrosine motifs within cytokine receptors. Additionally, upon phosphorylation of a tyrosine residue by a tyrosine kinase such as Janus kinase, the SH2 domain acts as a point for dimerization of two STAT proteins, either a heterodimer or homodimer [5]. Once STAT proteins dimerize, they move into the nucleus where they bind the gamma activated sequences (GAS) to initiate gene transcription. The TAD encodes a conserved serine phosphorylation site through which the protein interacts with co-factors/activators of transcription. In addition, the domain regulates protein stability, especially in STATs 4, 5, and 6; but not STAT1, 2, and 3, wherein sequences can be targeted by the ubiquitin–proteosome degradation pathway [3][6].

Figure 1. Protein/domain structure of STAT family members.
STAT Alternative Isoforms
In addition to full-length STATs, also termed STATα, alternative splicing and proteosomal cleavage results in the formation of STATβ and STATγ isoforms, respectively, as shown in Figure 2 [1]. STATβ protein possesses a modified 55 amino acid C-terminal that lacks the TAD. This impacts the protein’s ability to facilitate target gene transcription [7][8]. Studies have also demonstrated that overexpression of STATβ proteins may result in the protein having a dominant negative function. However, under physiological conditions, the two isoforms may also encode unique functions; for example, with STAT3, mice that only express the Stat3α isoform appear to have a defective response to endotoxic shock [9]. On the other hand, while STAT3 knockout is embryonic lethal, this can be rescued by expressing the STAT3β isoform [10]. STAT4 also has two isoforms, a full-length Stat4α or a truncated Stat4β lacking 44 amino acids in the C-terminus. IL-12 is a key cytokine that activates the expression of these two isoforms of STAT4. Microarray-based studies demonstrated that while both STAT4 isoforms activate 98 genes, Stat4α and Stat4β uniquely activated 32 and 29 genes, respectively [11]. What this suggests is that the β-isoforms of STAT3 and STAT4 possess transcription-inducing activity, further suggesting that these isoforms do not just possess a dominant-negative function. This further suggests that STAT-mediated signaling in response to cytokine activation is coordinated between the variants of the protein. Another method of regulating STAT proteins occurs in conditions without alternative splicing, but rather through proteolytic cleavage of the C-terminus. This eliminates the TAD, resulting in the STATβ isoform. Although STAT3, STAT5a, STAT5b, and STAT6 proteins form the γ-isoform, they each do so in different contexts. Located within the nucleus of myeloid progenitor cells, proteases cleave STAT5a and STAT5b to induce STAT5γ formation. This process is not affected by the tyrosine-phosphorylation state of STAT5 [1]. Just as with the cleavage of STAT5, in myeloid progenitor cells, proteases modify the C-terminal of STAT3α. However, this protease only cleaves the phosphorylated form of the full-length STAT3α. This phosphorylation requirement is a key difference between Stat3α and STAT5a/b. Such proteolytic processes occur primarily due to serine proteases, though calpain also demonstrates an affinity for cleaving STAT3 and STAT5 in platelets while cleaving STAT6 in mast cells [1].
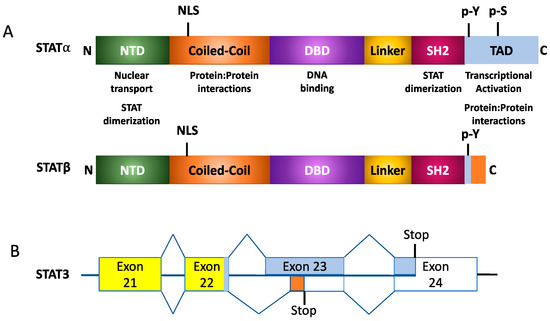
Figure 2. Structural organization of STAT isoforms. (A) Generalized protein organization of alpha and beta STAT isoforms following alternative splicing. (B) Example of splicing for STAT3α and STAT3β isoforms.
2. STAT Activation and Signaling
In the cytoplasm, STAT proteins exist in their inactive form before phosphorylation occurs, resulting in an active dimer that can translocate into the nucleus and bind to target DNA sequences and activate transcription [12]. A variety of signaling molecules and kinases are responsible for the activation of STAT proteins, but the primary and most well-known is the JAK/STAT pathway (Figure 3). Canonical signaling via JAK, and non-canonical activation of STATs are explored in detail in a recent review by Hu et al. [2]. Briefly, Janus kinase (JAK) is often activated by a transmembrane cytokine receptor [4]. The type of JAK and subsequent STAT activated depends on the cytokine and the receptor it activates, summarized in Table 1. Class I cytokine receptors respond to a wide range of cytokines including IL-6 and IL-2 that activate STAT3, STAT5, and STAT6. Interferon-γ activates STAT1, while interferon types II and III activate both STAT1 and STAT2. IL-10 cytokine activates STAT3 [13][14]. All four Janus kinases (JAK)—JAK1, JAK2, JAK3, and TYK2 (tyrosine kinase 2)—are activated by autophosphorylation after binding to the transmembrane receptor. STATs are then activated by phosphorylation by JAKs [15]. Canonical activation via JAK-induced stimuli results in phosphorylation of tyrosine residues within the TAD domain, promoting STAT dimerization, and nuclear import. STATs can also be activated at serine residues within the TAD, though this phosphorylation event remains inconclusive regarding function. Several studies have demonstrated that phosphorylation at these serine residues can enhance transcriptional activity, potentiating the effects of tyrosine phosphorylation [16][17][18]; however, it has also been reported to suppress transcriptional activity, further demonstrating the complexity of this signaling pathway [19][20][21]. Moreover, serine phosphorylation may therefore contribute to modulating transcriptional activity in response to select activators, regulating transcription of select target genes. These phosphorylation sites are summarized in Figure 1. There are a variety of other kinases that do not require a receptor and can activate STAT proteins without JAK. SRC is an example of a kinase that does not require a receptor [22]. Activation of STAT allows translocation from the cytoplasm into the nucleus, where it activates several genes related to development and survival of the cell [23].

Figure 3. STAT signaling. Cytokines, growth factors, and hormones bind to numerous receptors that result in the recruitment of JAK proteins, which, in turn, phosphorylate STAT proteins. Phosphorylated STATs sequester away from the receptor complex, dimerize, and translocate to the nucleus to enhance downstream target gene transcription.
Table 1. Activators of STAT signaling.
| Proteins | Activators | Biologic Response | References |
|---|---|---|---|
| STAT1 | IFNα, IFNβ, IFNγ, IFNλ, EGF, PDGF, IL6, IL27 | Promote inflammation, PD-L1 expression, apoptosis, MYC gene expression, monocyte activation | [24][25][26][27][28] |
| STAT2 | IFNα, IFNβ, IFNλ | Promote inflammation | [29] |
| STAT3 | IL6, IL6 family cytokines, IL10, IL22, Prolactin, Growth Hormone, EGF, TGFα, VEGF | Promote proliferation, chemoresistance, cell migration | [30] |
| STAT4 | IFNγ, IL12, IL23, IL2, IL35 | Promote T Cell differentiation, IFNγ production, NK cell activation | [31] |
| STAT5 | Growth hormone, Prolactin, IFNα, IFNβ, IL3, Thrombopoietin, Erythropoietin, GM-CSF | Promote proliferation, cell migration | [32][33] |
| STAT6 | IL4, IL13 | Promote macrophage polarization, EMT | [34][35] |
This entry is adapted from the peer-reviewed paper 10.3390/cancers15092485
References
- Hendry, L.; John, S. Regulation of STAT signalling by proteolytic processing. JBIC J. Biol. Inorg. Chem. 2004, 271, 4613–4620.
- Hu, X.; Li, J.; Fu, M.; Zhao, X.; Wang, W. The JAK/STAT signaling pathway: From bench to clinic. Signal Transduct. Target. Ther. 2021, 6, 402.
- Lim, C.P.; Cao, X. Structure, function, and regulation of STAT proteins. Mol. Biosyst. 2006, 2, 536–550.
- Pellegrini, S.; Dusanter-Fourt, I. The structure, regulation and function of the Janus kinases (JAKs) and the signal transducers and activators of transcription (STATs). JBIC J. Biol. Inorg. Chem. 1997, 248, 615–633.
- Heinrich, P.C.; Behrmann, I.; Müller-Newen, G.; Schaper, F.; Graeve, L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 1998, 334, 297–314.
- Schindler, C.; Plumlee, C. Inteferons pen the JAK–STAT pathway. Semin. Cell Dev. Biol. 2008, 19, 311–318.
- Caldenhoven, E.; van Dijk, T.B.; Solari, R.; Armstrong, J.; Raaijmakers, J.A.M.; Lammers, J.-W.J.; Koenderman, L.; de Groot, R.P. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 1996, 271, 13221–13227.
- Shao, H.; Quintero, A.J.; Tweardy, D.J. Identification and characterization of cis elements in the STAT3 gene regulating STAT3alpha and STAT3beta messenger RNA splicing. Blood 2001, 98, 3853–3856.
- Yoo, J.-Y.; Huso, D.L.; Nathans, D.; Desiderio, S. Specific ablation of Stat3beta distorts the pattern of Stat3-responsive gene expression and impairs recovery from endotoxic shock. Cell 2002, 108, 331–344.
- Maritano, D.; Sugrue, M.L.; Tininini, S.; Dewilde, S.; Strobl, B.; Fu, X.; Murray-Tait, V.; Chiarle, R.; Poli, V. The STAT3 isoforms alpha and beta have unique and specific functions. Nat. Immunol. 2004, 5, 401–409.
- Hoey, T.; Zhang, S.; Schmidt, N.; Yu, Q.; Ramchandani, S.; Xu, X.; Naeger, L.K.; Sun, Y.; Kaplan, M.H. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 2003, 22, 4237–4248.
- Ivashkiv, L.B.; Hu, X. Signaling by STATs. Arthritis Res. Ther. 2004, 6, 159–168.
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984–2009.
- Mowen, K.; David, M. Role of the STAT1-SH2 domain and STAT2 in the activation and nuclear translocation of STAT1. J. Biol. Chem. 1998, 273, 30073–30076.
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283.
- Wen, Z.; Zhong, Z.; Darnell, J.E., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 1995, 82, 241–250.
- Goh, K.C.; Haque, S.; Williams, B. P38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 1999, 18, 5601–5608.
- Kovarik, P.; Stoiber, D.; Novy, M.; Decker, T. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. EMBO J. 1998, 17, 3660–3668.
- Jain, N.; Zhang, T.; Fong, S.L.; Lim, C.P.; Cao, X. Repression of Stat3 activity by activation of mitogen-activated protein kinase (MAPK). Oncogene 1998, 17, 3157–3167.
- Lim, C.P.; Cao, X. Serine phosphorylation and negative regulation of Stat3 by JNK. J. Biol. Chem. 1999, 274, 31055–31061.
- Sengupta, T.K.; Talbot, E.S.; Scherle, P.A.; Ivashkiv, L.B. Rapid inhibition of interleukin-6 signaling and Stat3 activation mediated by mitogen-activated protein kinases. Proc. Natl. Acad. Sci. USA 1998, 95, 11107–11112.
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809.
- Gilmour, K.C.; Reich, N.C. Signal transduction and activation of gene transcription by interferons. Gene Expr. 1995, 5, 1–18.
- Tassiulas, I.; Hu, X.; Ho, H.; Kashyap, Y.; Paik, P.; Hu, Y.; Lowell, C.A.; Ivashkiv, L.B. Amplification of IFN-alpha-induced STAT1 activation and inflammatory function by Syk and ITAM-containing adaptors. Nat. Immunol. 2004, 5, 1181–1189.
- Cheng, C.; Lin, H.; Tsai, K.; Chiang, Y.; Lim, K.; Chen, C.G.; Su, Y.; Peng, C.; Ho, A.; Huang, L.; et al. Epidermal growth factor induces STAT1 expression to exacerbate the IFNr-mediated PD-L1 axis in epidermal growth factor receptor-positive cancers. Mol. Carcinog. 2018, 57, 1588–1598.
- Costa-Pereira, A.P.; Tininini, S.; Strobl, B.; Alonzi, T.; Schlaak, J.F.; Is’harc, H.; Gesualdo, I.; Newman, S.J.; Kerr, I.M.; Poli, V. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc. Natl. Acad. Sci. USA 2002, 99, 8043–8047.
- Kalliolias, G.D.; Ivashkiv, L.B. IL-27 activates human monocytes via STAT1 and suppresses IL-10 production but the inflammatory functions of IL-27 are abrogated by TLRs and p38. J. Immunol. 2008, 180, 6325–6333.
- Zhang, Y.; Liu, Z. STAT1 in cancer: Friend or foe? Discov. Med. 2017, 24, 19–29.
- Leung, S.; Qureshi, S.A.; Kerr, I.M.; Darnell, J.E.; Stark, G.R. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell Biol. 1995, 15, 1312–1317.
- Lu, W.; Chen, H.; Yel, F.; Wang, F.; Xie, X. VEGF induces phosphorylation of STAT3 through binding VEGFR2 in ovarian carcinoma cells in vitro. Eur. J. Gynaecol. Oncol. 2006, 27, 363–369.
- Morinobu, A.; Gadina, M.; Strober, W.; Visconti, R.; Fornace, A.; Montagna, C.; Feldman, G.M.; Nishikomori, R.; O’Shea, J.J. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc. Natl. Acad. Sci. USA 2002, 99, 12281–12286.
- Alkharusi, A.; AlMuslahi, A.; AlBalushi, N.; AlAjmi, R.; AlRawahi, S.; AlFarqani, A.; Norstedt, G.; Zadjali, F. Connections between prolactin and ovarian cancer. PLoS ONE 2021, 16, e0255701.
- Tsuji-Takayama, K.; Otani, T.; Inoue, T.; Nakamura, S.; Motoda, R.; Kibata, M.; Orita, K. Erythropoietin induces sustained phosphorylation of STAT5 in primitive but not definitive erythrocytes generated from mouse embryonic stem cells. Exp. Hematol. 2006, 34, 1323–1332.
- Waqas, S.F.H.; Ampem, G.; Röszer, T. Analysis of IL-4/STAT6 Signaling in Macrophages. Methods Mol. Biol. 2019, 1966, 211–224.
- Cao, H.; Zhang, J.; Liu, H.; Wan, L.; Zhang, H.; Huang, Q.; Xu, E.; Lai, M. IL-13/STAT6 signaling plays a critical role in the epithelial-mesenchymal transition of colorectal cancer cells. Oncotarget 2016, 7, 61183–61198.
This entry is offline, you can click here to edit this entry!
