The microRNAs (miRNAs), lncRNAs (long ncRNAs), and circRNAs (circular RNAs) with significant regulatory and structural roles make up approximately 99% of the human genome, which does not contain proteins. Non-coding RNAs (ncRNA) have been discovered to be essential novel regulators of cardiovascular risk factors and cellular processes, making them significant prospects for advanced diagnostics and prognosis evaluation. Cases of CVDs are rising due to limitations in the current therapeutic approach; most of the treatment options are based on the coding transcripts that encode proteins. Recently, various investigations have shown the role of nc-RNA in the early diagnosis and treatment of CVDs. Furthermore, the development of novel diagnoses and treatments based on miRNAs, lncRNAs, and circRNAs could be more helpful in the clinical management of patients with CVDs. CVDs are classified into various types of heart diseases, including cardiac hypertrophy (CH), heart failure (HF), rheumatic heart disease (RHD), acute coronary syndrome (ACS), myocardial infarction (MI), atherosclerosis (AS), myocardial fibrosis (MF), arrhythmia (ARR), and pulmonary arterial hypertension (PAH). Here, we discuss the biological and clinical importance of miRNAs, lncRNAs, and circRNAs and their expression profiles and manipulation of non-coding transcripts in CVDs, which will deliver an in-depth knowledge of the role of ncRNAs in CVDs for progressing new clinical diagnosis and treatment.
- cardiovascular disease
- microRNAs
- diagnosis
- long noncoding RNA
- ncRNAs
- therapy
1. Introduction
2. miRNAs and CVDs
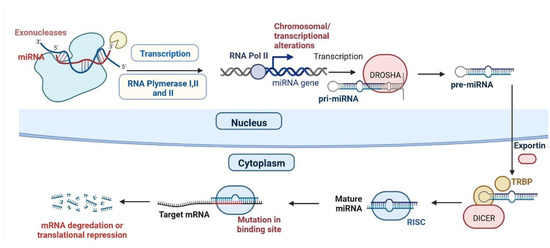
| S.N. | Type of Disease | miRNA | Regulation | Importance | Reference |
|---|---|---|---|---|---|
| 1 | Cardiac hypertrophy (CH) | miR-208a | Up-regulation | Cardiac remodelling | [13] |
| 2 | CH | miR-19a/b | Up-regulation | Cardiac remodelling in response to angiotensin II infusion | [14] |
| 3 | CH | miR-155 | Up-regulation | Cardiac remodelling | [15] |
| 4 | CH | miR-199a | Up-regulation | Maintenance of cell size in cardiomyocytes | [16] |
| 5 | CH | miR-1, | Down-regulation | Induces cardiac hypertrophy. | [17] |
| 6 | CH | miR-101 | Down-regulation | Inhibit cardiac hypertrophy signalling | [18] |
| 7 | CH | miR-185 | Down-regulation | Inhibit CH hypertrophy signalling | [19] |
| 8 | CH | miR-34a | Down-regulation | Regulation of Ang II-induced cardi myocyte hypertrophy | [20] |
| 9 | CH | miR-145 | Down-regulation | Inhibits isoproterenol-induced cardiomyocyte hypertrophy | [21] |
| 10 | CH | miR-150 | Down-regulation | Reduces the immunosuppression function of Myeloid-derived suppressor cells (MDSCs) | [22] |
| 11 | CH | miR-378 | Down-regulation | Act as negative regulator for CH | [23] |
| 12 | Heart failure (HF) | miR-125b | Up-regulation | Conduction of Cardiac fibrosis (CF) | [24] |
| 13 | HF | miR-22, | Up-regulation | Regulator for cardiac remodelling | |
| 14 | HF | miR-92b | Up-regulation | Related to the left atrium diameter, left ventricular end-diastolic dimension | [25] |
| 15 | HF | miR-320a | Up-regulation | CF through activation of the IL6/STAT3 axis. | [26] |
| 16 | HF | miR-423-5p | Up-regulation | Upregulated in human failing myocardium | [27] |
| 17 | HF | miR-200b | Up-regulation | Regulation of multiple cellular pathways in HF | [28] |
| 18 | HF | miR-622 | Up-regulation | Improves blood vessel growth | [29] |
| 19 | HF | miR-1228 | Up-regulation | Marker for systolic HF | [30] |
| 20 | HF | miR-208b | Up-regulation | pathogenesis of DCM | [31] |
| 21 | HF | miR-499 | Up-regulation | Cardiac development | [32] |
| 22 | HF | miR-223 | Up-regulation | Altered in post-MI HF in humans | [33] |
| 23 | HF | miR-1254 | Up-regulation | Altered in post-MI HF in humans | [34] |
| 24 | HF | miR-1306 | Up-regulation | Elected to explore novel circulating markers for HF | [35] |
| 25 | HF | miR-18a | Down-regulation | CF through the Notch2 pathway. | [36] |
| 26 | HF | miR-26b | Down-regulation | Controlling critical signalling pathways, such as BMP/ SMAD1 signalling | [37] |
| 27 | HF | miR-27a | Down-regulation | Inhibiting miR-27a-3p mitigated CH phenotype induced by Ang II (Angiotensin -II) | [38] |
| 28 | HF | miR-30e | Down-regulation | The overexpression of miR-30c reduces the level of connective tissue growth | [39] |
| 29 | HF | miR-106a | Down-regulation | Notch 3 pathway in ischemic heart injury. | [40] |
| 30 | HF | miR-199a | Down-regulation | Improves contractile function | [41] |
| 31 | HF | miR-652 | Down-regulation | Marker for predicting acute coronary syndrome | [42] |
| 32 | HF | miR-1 | Down-regulation | Systolic HF | [43] |
| 33 | HF | miR-126 | Down-regulation | Activation of the vascular endothelial growth factor (VEGM) signalling pathway in the endothelium. | [44] |
| 34 | HF | miR-423 | Down-regulation | It is a circulating biomarker for heart failure. | [45] |
| 35 | Cardiac electrical and structural remodelling (CE and SR) | miR-1 | Down-regulated | Increased altered conduction Increased CF |
[46] |
| 36 | CE and SR | miR-26 | Down-regulated | Increase inwardly rectifying channel | [47] |
| 37 | CE and SR | miR-29 | Down-regulated | Increased CF | [48] |
| 38 | CE and SR | miR-30 | Down-regulated | Increased CF | [49] |
| 39 | CE and SR | miR-133 | Down-regulated | Increased CF | [50] |
| 40 | CE and SR | miR-328 | Up-regulated | Shortened atrial action potential duration by targeting | [51] |
| 41 | CE and SR | miR-499 | Up-regulated | Altered conduction by targeting |
[52] |
| 42 | CE and SR | miR-21 | Up-regulated | Inhibition of fibroblast proliferation | [53] |
| 43 | Acute coronary syndrome (ACS) and myocardial infarction (MI) | miR-1 | Up-regulated | marker of cardiomyocyte injury | [54] |
| 44 | ACS and MI | miR-133a | Up-regulated | Development of VF (Ventricular fibrillation) | [55] |
| 45 | ACS and MI | miR-208a | Up-regulated | Regulates the cardiac stress response. | [56] |
| 46 | ACS and MI | miR-499-5p | Up-regulated | Associated with cardiac injury and also with cardio protection | [57] |
| 47 | ACS and MI | miR-126, | Down-regulated | downregulated in the region adjacent to MI areas | [58] |
| 48 | ACS and MI | miR-221/222 | Down-regulated | Severity of the coronary artery lesions | [59] |
| 49 | ACS and MI | miR-29 | Dysregulation | Involved in CF multiple collagens, fibrillin’s, and elastin | [60] |
| 50 | ACS and MI | miR-145 | Up-regulated | Significantly upregulated in mice in response to chronic hypoxia and that genetic ablation | [61] |
| 51 | ACS and MI | miR-21 | Up-regulated | Up-regulated in the hypoxia | [62] |
| 52 | ACS and MI | miR-206 | Up-regulated | Normal and hypertensive mouse PASMCs. | [63] |
| 53 | ACS and MI | miR-328 | Down-regulated | Regulates Hypoxic Pulmonary Hypertension | [64] |
| 54 | Pulmonary arterial hypertension (PAH) | miR-204 | Down-regulated | Hypoxia related to pulmonary arterial hypertension | [65] |
| 55 | Atherosclerosis (AS) | miR-33 | Dysregulation | Promising strategy to reverse autophagy dysfunction in atherosclerosis. | [66] |
| 56 | (AS) | miR-122 | Up | Significantly up-regulated in patients with atherosclerotic lesion | [67] |
| 57 | (AS) | miR-126 | Down-regulated | [68] | |
| 58 | (AS) | miR-1 | Down-regulated | Downregulation of miR-10a enhances IκB/NF-κB activation | [69] |
| 59 | (AS) | miR-221/222 | Down-regulated | Suppression of PGC-1α (peroxisome proliferator-activated receptor gamma coactivator 1-alpha) in the progression of atherosclerosis | [70] |
| 60 | Congenital heart diseases (CHDs) | miR-1275, miR-27b, miR-421 | Up-regulated | usually developing hearts | [13] |
| 61 | CHD | miR-122, miR-1201 | Down-regulated | developing hearts | [14] |
| 62 | CHD | miR-222, miR-337-5p, miR-363, miR-424, miR-424, miR-660, miR-708, miR-421, miR-19a, miR-130b, miR-146b-5p, miR-154, miR-155, miR-181c, miR-181d and miR-192, | Up-regulated | tetralogy of Fallot | [15] |
| 63 | CHDs | miR-181a, miR-720, miR-29c and miR-940 | Down-regulated | tetralogy of Fallot | [16] |
| 64 | CHD | miR-181c | Up-regulated | ventricular septal defect | [17] |
| CHD | miR-1-1 | Down-regulated | ventricular septal defect | [18] | |
| 65 | CHD | miR-106a, miR-144, miR-451, miR-486-3p, miR-486-5p, hsa-let-7e, miR-16, miR-18a, miR-25, miR-93, and miR-505 | Up-regulated | transposition of the great arteries | [19] |
| 66 | CHD | miR-873 | Up-regulated | Cyanotic CHD | [20] |
| 67 | CHD | miR-182 | Down-regulated | Cyanotic CHD | [21] |
| 68 | CHD | miR-498 | Up-regulated | Ventricular septal defect | |
| 69 | CHD | miR-379-5p, miR-409-3p, miR-433, hsa-let-7e-5p, miR-155-5p, miR-222-3p, and miR-487b | Down-regulated | Ventricular septal defect | [22] |
| 70 | CHD | hsa-let-7b, hsa-let-7a, and miR-486 | Up-regulated | Atrioventricular septal defect and atrial septal defect | [23] |
| 71 | CHD | miR-19b, miR-22, miR-29c, miR-375 | Up-regulated | Atrioventricular septal defect and atrial septal defect | [22,23] |
| 72 | Myocrdial infraction (I) | miR-1 | Cardiomyocyte Downstream Targets: Ncx-1; KCNJ2, GJA1; IGF-1 |
[55] | |
| 73 | MI | miR-15 | Up-regulated | Cardiomyocyte Downstream Targets: Pdk4, Sgk1 |
[56] |
| 74 | MI | miR-21 | Down-regulated | Fibroblast, Downstream Targets Pten; Sprouty-1, collagens | [57] |
| 75 | MI | miR-24 | Up-regulated | Anti-apoptosis in Cardiomyocyte, fibroblast, endothelial cell; Downstream Targets Bim; Furin; Gata2, Pak4 |
[58] |
| 76 | MI | miR-29 | Down-regulated | Cardiomyocyte, fibroblast Downstream Targets: Mcl-1; Collagens |
[59] |
| 77 | MI | miR-92a | Up-regulated | Endothelial cell Downstream Targets: Itga5 |
[60] |
| 78 | MI | miR-101 | Down-regulated | Cardiac remodelling Downstream Targets: Collagens |
[61] |
| 79 | MI | miR-126 | Down-regulated | Protects against myocardial ischemia-reperfusion injury | [62] |
2.1. miRNAs and HF
2.2. Arrhythmias
2.3. miRNAs and ACS and MI
2.4. miRNAs and Atherosclerosis
2.5. miRNAs and RHD
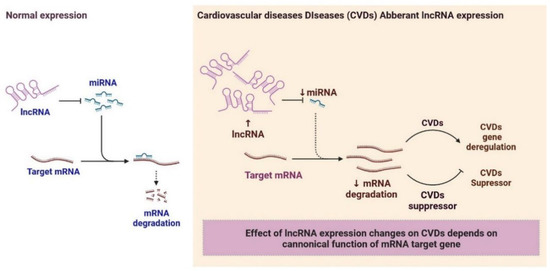
2.6. LncRNAs and Cardiovascular Diseases
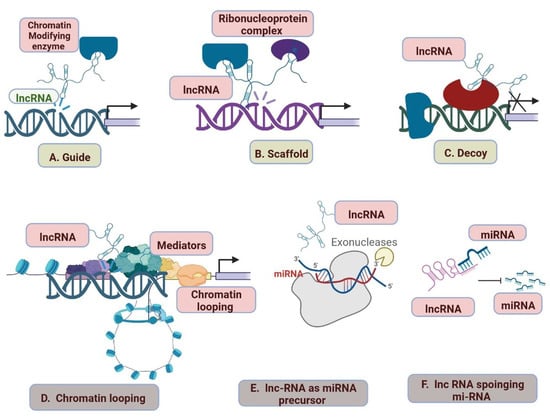
| Type of Disease | lncRNA | Regulations | Importance | References | |
|---|---|---|---|---|---|
| 1 | Myocardial infraction (MI) | aHIF | Regulations of the angiogenesis process and a biomarker | Inhibits the autophagy of cardiac cells during MI | [102] |
| 2 | MI | ANRIL | Regulates myocardial cell apoptosis in AMI | Protection of cardiomyocytes | [103] |
| 3 | MI | APF | APF lncRNA regulates autophagy | Acting as a sponge for miRNA- 188-3p. |
[102] |
| 4 | MI | CARL | Regulates mitochondrial fission and apoptosis | Acting as a sponge for miRNA-539. |
[102] |
| 5 | MI | CDR1AS | Inhibiting the autophagy of cardiac cells during MI | Biomarker. | [102] |
| 6 | MI | FTX | Regulates cardiomyocytes | Act as a sponge for miRNA- 29b-1-5. |
[104] |
| 7 | MI | GAS5 | Regulates the protection of cardiomyocytes against hypoxic injury | Act as a sponge for miRNA-142; improves apoptosis by negatively regulating sema3a. |
[102] |
| 8 | MI | H19 | Regulates autophagy | Induction of cardiac remodeling, autophagy, and biomarker. | [102] |
| 9 | MI | HOTAIR | Regulates cardioprotective | Act as a sponge for miRNA-1 and as a biomarker. |
[105] |
| 10 | MI | KCNQ1OT1 | Down-regulation of lncRNA KCNQ1OT1 protects against myocardial ischemia/reperfusion injury | Biomarker for left ventricular dysfunction. | [106] |
| 11 | MI | LIPCAR | Down-regulated | Biomarker for cardiac remodelling. | [107] |
| 12 | MI | Lnc-Ang362 | Upregulation | Promotion of CF | [108] |
| MI | MALAT1 | Down-regulation | Regulation of cardiomyocytes apoptosis and autophagy through miRNA- 558; and biomarker. |
[109] | |
| 13 | MI | MDRL | Regulates mitochondrial fission | Reduction of mitochondrial fission and apoptosis acting as a sponge for miRNA-361. |
[110] |
| 14 | MI | MEG3 | Regulates cardiomyocytes | Regulation of cardiomyocytes apoptosis. | [111] |
| 15 | MI | MHRT | Regulates cardiomyocytes | Regulation of cardiomyocytes apoptosis and biomarker. | [112] |
| 16 | MI | MIAT | Regulates CF | Regulation of cardiac hypertrophy and fibrosis acting as a sponge for miRNA-150 and -93. |
[102] |
| 17 | MI | Mirt1/2 | Regulates cardiomyocytes | Regulation of cardiac remodelling. | [102] |
| 18 | MI | n379519 | Regulates CF | Promotion of cardiac fibrosis through miRNA-30. | [113] |
| 19 | MI | NONRATT021972 | Regulates cardiomyocytes | Promotion of cardiac function. | [114] |
| 20 | MI | NRF | Regulates cardiomyocyte necrosis. | Regulation of cardiomyocyte necrosis. | [115] |
| 21 | MI | NRON | Up-regulated | Wisper in cardiac fibroblast; Biomarker | [102] |
| 22 | MI | PCFL | Up-regulated | Promotion of cardiac fibrosis through miRNA-378. | [102] |
| 23 | MI | TTTY15 | Up-regulated | Induction of cardiomyocyte injury by hypoxia targeting miRNA-455. | [102] |
| 24 | MI | UCA1 | Regulates cardiomyocytes | Regulated ischemia and hypoxia of cardiomyocytes; Biomarker. | [115] |
| 25 | MI | UIHTC | Regulates cardiomyocytes against MI | Promotion of mitochondrial function. | [102] |
| 26 | MI | Wisper | Regulates cardiac fibroblast | MI-induced fibrosis and cardiac dysfunction | [116] |
| 27 | MI | ZFAS1 | Regulates cardiomyocyte | Induction of cardiomyocyte apoptosis, cardiac contractility reduction, and biomarker. |
[117] |
| 28 | Coronary heart disease | aHIF | Up-regulated | Biomarker. | [118] |
| 29 | Coronary heart disease | ANRIL | Down-regulates | Diagnostic and prognostic indicator for CHD | [118] |
| 30 | Coronary heart disease | APOA1-AS | Up-regulations increase the risk of CHD | Biomarker. | [119] |
| 31 | Coronary heart disease | AWPPH | Regulates apoptosis. | Promotion of ECs apoptosis. | [120] |
| 32 | Coronary heart disease | BACE1-AS | dysregulation | dysregulation of the BACE1/BACE1-AS/Aβ axis is associated with HF. | [121] |
| 33 | Coronary heart disease | BANCR | Differentially expressed | Promotion of VSMCs proliferation and migration. | [122] |
| 34 | Coronary heart disease | CHROME | Up-regulated | Regulation of cellular cholesterol homeostasis | [123] |
| 35 | Coronary heart disease | CoroMarker | Differentially expressed | novel biomarker for the diagnosis | [124] |
| 36 | Coronary heart disease | EGOT | Differentially expressed | Biomarker. | [125] |
| 37 | Coronary heart disease | H19 | Differentially expressed | Biomarker. | [126] |
| 38 | Coronary heart disease | HOTTIP | Up-regulates | Promotes ECs proliferation and migration | [127] |
| 39 | Coronary heart disease | HRCR | Regulates hypertrophic Ca2+ signaling pathway | Regulation of cardiomyocytes apoptosis and proliferation. | [128] |
| 40 | Coronary heart disease | LIPCAR | Differentially expressed | Biomarker. | [107] |
| 41 | Coronary heart disease | lincRNA-p21 | Regulates cardiac remodelling and heart failure | Regulation of cardiomyocytes apoptosis and proliferation. | [129] |
| 42 | Coronary heart disease | LINC00968 | Up-regulated | Promotion of ECs proliferation and migration acting as a sponge for miRNA-9 |
[130] |
| 43 | Coronary heart disease | MALAT1 | Differentially expressed | Biomarker. | [131] |
| 44 | Coronary heart disease | MIAT | Differentially expressed | Biomarker | [132] |
| 45 | Coronary heart disease | NEXN-AS1 | Differentially expressed | Mitigation of atherosclerosis. | [133] |
| 46 | Coronary heart disease | SMILR | Differentially expressed | Biomarker. | [134] |
| 47 | Arterial Hypertension | AK098656 | Up-regulated | Regulation of arteries of resistance and a biomarker | [135] |
| 48 | Arterial Hypertension | ANRIL | Regulates endothelial cell activities | Increase of susceptibility to higher systolic blood pressure conferred by polymorphisms. |
[136] |
| 49 | Arterial Hypertension | GAS5 | Regulates ECs and VSMCs function | Regulation of ECs and VSMCs function acting as endogenous RNA competing of miRNA-21; and a biomarker. GAS5 Targets miR-194-3p. miR-194-3 |
[137] |
| 50 | Arterial Hypertension | Giver | Regulates VSMCs dysfunction. | Promotion of VSMCs dysfunction. | [138] |
| 51 | Arterial Hypertension | Lnc-Ang362 | Regulates VSMCs | Regulation of VSMCs proliferation through miRNA-221 and -222. | [139] |
| 52 | Arterial Hypertension | NR_027032 | Differentially expressed | Biomarker. | [140] |
| 53 | Arterial Hypertension | NR_034083 | Differentially expressed | Biomarker. | [141] |
| 54 | Arterial Hypertension | NR_104181 | Differentially expressed | Biomarker. | [140] |
| 55 | Heart failure | ANRIL | Differentially expressed | Biomarker. | [142] |
| 56 | Heart failure | BACE1-AS | Regulates apoptosis. | Promotion of ECs apoptosis. | [143] |
| 57 | Heart failure | Chaer | Dysregulation | Induction of Pathological cardiac remodelling. | [144] |
| 58 | Heart failure | Chast | Down-regulates | Induction of Pathological cardiac remodelling. | [145] |
| 59 | Heart failure | CHRF | Up-regulated | Endogenous sponge to miRNA-489 activity. | [146] |
| 60 | Heart failure | HEAT2 | Up-regulated | Biomarker. | [147] |
| 61 | Heart failure | HOTAIR | Up-regulated | LncRNA HOTAIR may function as a miR-19-sponge to modulate PTEN levels Biomarker. | [148] |
| 62 | Heart failure | LIPCAR | Up-regulated | Biomarker. | [149] |
| 63 | Heart failure | lincRNA-ROR | Regulates CH | Regulation of cardiac hypertrophy acting as a sponge for miRNA-133. | [150] |
| 64 | Heart failure | LOC285194 | Up-regulated | overexpression suppressed MKN45 and HGC-27 cell proliferation and promoted cell apoptosis; Biomarker. | [148] |
| 65 | Heart failure | MEG3 | Regulates CF | Regulation of cardiac fibrosis and diastolic dysfunction | [151] |
| 66 | Heart failure | MHRT | Regulates of chromatin re-modellers | Regulation of chromatin remodels and biomarker. | [152] |
| 67 | Heart failure | MIAT | Regulates CH | Regulation of cardiac hypertrophy acting as a sponge for miRNA-150. | [153] |
| 68 | Heart failure | NRON | Upregulated | Biomarker. | [154] |
| 69 | Heart failure | RNY5 | Dysregulation | Biomarker. | [155] |
| 70 | Heart failure | SOX2-OT | Dysregulation | Biomarker. | [156] |
| 71 | Heart failure | SRA1 | Dysregulation | Biomarker. | [157] |
This entry is adapted from the peer-reviewed paper 10.3390/cells12121629
