1. Autophagy and AML Stem Cells
During leukemic development, LSCs can adapt their metabolism and autophagic mechanisms to provide the high energy and nutrients required for LSC proliferation and survival under conditions of nutrient deficiency, starvation, hypoxia, or during chemotherapy treatments [7
, 91
, 92
, 93
, 94
] .
Mitophagy, mitochondrial function and integrity can affect the viability, proliferation and differentiation potential and longevity of normal hematopoietic stem cells (HSCs) [95] and are important in the survival strategy
of AML stem cells (LSCs) .
Specifically, adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK), a protein complex critical in mitochondrial metabolism and mitophagy, is constitutively activated in LSCs, increasing mitochondrial clearance to support LSC growth and survival through its downstream target FIS1, the mitochondrial fission 1 protein component of a mitochondrial complex that promotes mitochondrial fission [
96 ]. AMPK/FIS1-mediated mitophagy is required for LSC self-renewal and survival [
96]. Overexpression of FIS1 was also found in AML cells while depletion of FIS1 impairs mitophagy, weakening the self-renewal capacity of LSCs and resulting in the induction of myeloid differentiation by inactivation of GSK3 (glycogen synthase kinase 3), thus pointing to mitophagy as a regulatory mechanism for AML progression [
96 ]. More recently, the loss of sequestosome 1 (
SQSTM1 ), also known as p62, a selective autophagy receptor crucial for the development and progression of AML in vivo, induces the accumulation of damaged mitochondria and mitochondrial superoxide, thereby compromising cell survival. leukemia cells. Thus, the loss of
SQSTM1impairs leukemia progression in mouse models of AML, underscoring the role of mitophagy in LSC survival.
97 ]. Collectively, these studies demonstrate that increased autophagic activity of LSCs is required for malignant progression into AML.
However, in contrast to the activation of autophagy observed in AML, a loss of autophagy in healthy HSCs triggers the expansion of a population of progenitor cells in the bone marrow, resulting in severe and invasive myeloproliferation, as in human AML [ 53
] . This apparent paradox may be explained by the distinct roles that autophagy may play during AML progression, which may be different at various stages of leukemogenesis [
98 ,
99]. Autophagy in normal HSCs may prevent the onset of cancer, as a tumor suppressive mechanism. Indeed, autophagy removes damaged organelles, such as mitochondria, and protects hematopoietic cells from genomic instability and inflammation, thus preventing the onset of leukemia. Notably, increased DNA damage, high ROS levels, aneuploidy, and an aberrant accumulation of p62/SQSTM1 have been correlated with an impaired autophagy process, indicating a key role of autophagy in preventing the initiation of tumor [100
] . Conversely, in established cancer, autophagy may function as a favorable pathway that promotes tumor survival and growth,
Some studies have also shown that the activation of the autophagic flux only plays a role in the initiation of AML, with a transformation from HSC to LSC, and, therefore, after this stage, autophagy is not required for disease maintenance [101
] . AML studies MLL-AF9, the most common alteration in childhood AML, point to ATG5 or ATG7 as required for the onset of AML, but once the leukemic condition is established, autophagy is not required for function LSC in vivo [101
, 102
] . However, in a different AML MLL-ENL mouse model, Atg5 or Atg7 knockout reduced the number of functional LSCs, increased the
. ]. In this context, during the process of leukemogenesis, histone methylation can regulate central autophagic effectors and upstream autophagic regulators such as ATG 5 and ATG7 to indirectly affect the level of autophagy [104
] . Together, these studies suggest a highly complex and context-dependent role for autophagy in leukemic transformation with respect to the maintenance properties of LSCs in AML.
The dual role of autophagy in AML, as a promoter or suppressor of cancer in AML, is still a matter of debate. Studies have demonstrated that autophagy can act as a pro- or anti-proliferative mechanism depending on the lineage and molecular genotypic context of the disease, reflecting the degree of heterogeneity of AML [105
] .
2. Regulation of autophagy genes in AML cells
Numerous studies have demonstrated that increased autophagy in AML cells confers protection from chemotherapy treatment and promotes AML cell survival.
Increased ATG7-mediated autophagy has been associated with poor clinical outcomes and short duration of remission in AML patients [
106 ]. More recently, some proteins involved in leukemia cell survival and overexpressed in AML have been related to ATG overexpression, underlying the interplay between autophagy and protein overexpression that promotes leukemia cell survival [8]
.]. Hu et al. demonstrated that elevated expression of SIRT1 (Sirtuin 1), a key player in mitochondrial biogenesis and autophagy-related protein, is associated with elevated expression of CXCR4, a negative prognostic marker in AML, and other proteins related to autophagy such as ATG5 and LC3 in primary human AML samples, indicating a potential role of the SDF-1α-CXCR4 signaling pathway in the induction of autophagy in AML cells, which further promotes their survival under stress [107
] .
The transient melastatin receptor 2 (TRPM2) ion channel, involved in the maintenance of cell survival after oxidative damage, is highly expressed in AML [
108 ].
By performing TRPM2 depletion , Chen SJ et al. [
108 ] demonstrated that the levels of ULK1, Atg7 and Atg5 proteins are decreased in
TRPM2- depleted cells , leading to inhibition of autophagy. Importantly,
TRPM2 depletion in AML inhibits leukemia proliferation and increases the doxorubicin sensitivity of AML cells [
108 ].
Functional studies in normal CD34+CB cells indicated that inhibition of VMP1 expression reduced autophagic flux, with decreased hematopoietic stem and progenitor cell (HSPC) expansion, delayed differentiation, increased apoptosis, and impaired cellular function and engraftment in vivo. Similar results were observed in leukemic cell lines and primary CD34+ AML cells. Furthermore, ultrastructural analysis indicated that leukemic cells overexpressing VMP1 have a reduced number of mitochondrial structures and the number of lysosomal degradation structures is increased. Overexpression of VMP1 (vacuole membrane protein-1) increased autophagic flux and improved mitochondrial quality,
109 ].
Heterozygous deletions, missense mutations, or changes in copy number of key autophagy genes have been found with a high frequency in AML patients, particularly in AML patients with complex karyotypes [5
, 103
] . In particular, a heterozygous chromosomal loss of 5q, 16q, or 17p correlates with regions encoding the autophagic genes
ATG10 and
ATG12 ,
GABARAPL2 , and
MAP1LC3B or
GABARAP , respectively [
103 ], and many other autophagic genes have a low level of expression in the Human AML blasts, a reduced autophagic flux, and high levels of ROS [
103]. Furthermore, one study suggested that key autophagy genes such as
ULK1 ,
ATG3 ,
ATG4D and
ATG5 were significantly downregulated in primary AML cells compared to normal granulocytes [
110 ].
3. Autophagic biomarkers
Significant progress has recently been made to identify specific autophagy-related genes for the prediction of clinical outcomes in AML.
Along with the previously described ATG genes , several microRNAs implicated in leukemogenesis and chemoresistance have also been involved in autophagy activation and can be used as biomarkers [
111 ]. Notably, miR-17-5p overexpression in leukemia promotes AML proliferation by inhibiting autophagy through BECN1 targeting [
112 ,
113 ,
114]. Gansan et al. demonstrated that stromal cells downregulate miR-23a-5p levels in leukemia cells, leading to upregulation of protective autophagy in these cells, thereby increasing their resistance to chemotherapy toxicity.
115 ]. Overexpression of MiR-143 has been shown to enhance the sensitivity of AML cells to the cytotoxicity of cytarabine (Ara-C) treatment by inhibiting autophagy through targeting ATG7 and ATG2B [116
] . An overexpression of miR-15a-5p is involved in the chemoresistance of AML patients, through the autophagy-related genes
ATG9A ,
ATG14 ,
GABARAPL1 and
SMPD1which target AML cells [
117 ].
Recent advances in bioinformatics have yielded an autophagy-related signature that may help predict overall survival (OS) and/or clinical outcomes of patients with AML. Several studies have shown that the progression of AML depends on the gene signature associated with autophagy [
118 ]. A recent bioinformatics study constructed a model containing 10 autophagy-related genes to predict survival of AML patients, demonstrating that groups at high AML risk have higher expression of immune checkpoint genes and a higher percentage of CD4 T and NK cells [119]
.]. Furthermore, this study was able to predict OS in AML through the signature of 10 genes, pointing to this model as an effective prognostic predictor for AML patients, useful to guide patient stratification for immunotherapies and medications [119]
. . The LASSO Cox bioinformatics regression study which identified a critical risk signature for AML, consisting of the autophagic genes
BAG3 ,
CALCOCO2 ,
CAMKK2 ,
CANX ,
DAPK1 ,
P4HB ,
TSC2 and
ULK1 , had excellent predictive power for AML prognosis [
120]. Notably, immunosuppressive cytokines were found to be significantly increased in the tumor microenvironment of patients with a high risk of AML, predicted on the basis of the autophagy-related signature of these patients [120
] . However, the prognostic value of the ATG signature in the clinical setting is still debated. Therefore, the roles of the ATG signature and autophagy in the pathogenesis of AML should be further investigated.
Furthermore, an interesting study indicated that an autophagy-related lncRNA signature containing six lncRNAs (
HYMAI ,
MIR155HG ,
MGC12916 ,
DIRC3 ,
C1orf220 and
HCP5 ) may have an important prognostic value [
121 ]. A recent study pointed to four autophagy-associated lncRNAs (
MIR133A1HG ,
AL359715.1 ,
MIRLET7BHG , and
AL356752.1 ) as signatures to potentially be used as a biomarker to predict survival of AML patients [
122 ].
Collectively, these data indicate that the role of autophagy in tumor development clearly depends on the type of AML and stage of tumor development. Furthermore, autophagy may provide cancer cells with a survival strategy, suggesting a therapeutic use for autophagy inhibition. On the other hand, autophagy can induce cell death, pointing to autophagy activation as a novel strategy in cancer therapy. Therefore, it is necessary to determine the role played by autophagy in the molecular subtypes of AML, or the degree of tumor development, to verify whether its modulation could lead to benefits for the treated patient.
4. Autophagy and genetic alterations in AML
The AML phenotype results from multiple molecular, genetic, and epigenetic alterations that affect the differentiation, proliferation, and apoptosis of myeloid progenitors. The World Health Organization has classified AMLs based on the presence of particular genetic alterations, often originating from chromosomal translocations or other genome rearrangements such as t(8;21), t(15;17), inv(16), inv (3) , t(6;9), t(9;11) or t(11;19), or mutations in receptor kinases, key signaling mediators, proto-oncogenes, or epigenetic enzymes, for example, mutations in FLT3 (FMS
- like tyrosine kinase 3),
TP53 ,
c-KIT or
IDH1/2 ,
NPM1(nucleophosmin 1) and CCAAT enhancer binding protein (
CEBPA ) [
1 ,
2 ,
123 ]. These mutations in AML have an impact on the choice of the most suitable therapy.
The association between autophagy and recurrent genetic alterations has been described in several AML studies, but needs further investigation [
124 ,
125 ]. Here, we summarize and update recent advances that have highlighted the link between autophagy and fusion genes and recurrent oncogenic mutations in AML and the involvement of autophagy in chemotherapy treatment (
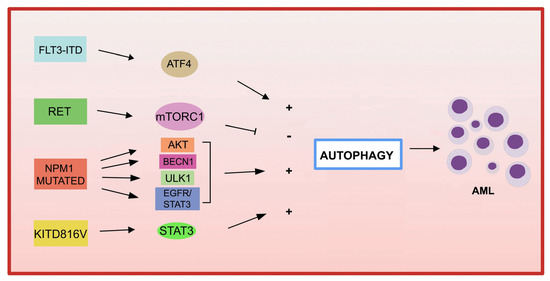
Figure 1 ).
Figure 1. Gene mutations in AML and autophagy. Several recurrent genetic abnormalities in AML are involved in the deregulation of autophagy, leading to leukemia progression.
4.1. Fusion genes in AML and autophagy
Most cases of APL are caused by a de novo translocation t(15;17)(q22;q21), resulting in the fusion of the
RARA gene with the
PML gene [
126 ,
127 ]. APL cells that have lower expression of autophagy-related genes than normal cells have reduced autophagic activity.
Using differentiating agents, such as all- trans- retinoid acid (ATRA) and arsenic trioxide (ATO) currently used in clinical settings, the expression level of autophagy-related genes increases, thus restoring autophagy in APL cells [
128]. Both agents can activate ETosis, a type of cell death mediated by the release of neutrophil extracellular traps (ET). Furthermore, mTOR-dependent autophagic action is required for ATO-induced NETosis in APL cells [
129 ]. Of note, rapamycin, the inhibitor of the mTOR pathway, synergizes with ATO in the eradication of leukemia-initiating cells (LIC) through the activation of NETosis in both APL cells and an in vivo APL model [129
] .
Translocations of the mixed lineage leukemia (
MLL ) 11q23 gene are observed in approximately 80% of pediatric AMLs.
In these, the MLL gene can, by genomic translocation, be fused with >60 different fusion partners [
130 ]. Treatment with the RAS oncogene inhibitor, tipifamib, leads to inhibition of AML by the t(6;11) translocation inducing both apoptosis and autophagy [
131 ]. Another study demonstrated that ATG5 participates in the development of
MLL-AF9- driven leukemia , but not in AML-sensitive chemotherapy mice expressing MLL-AF9 [
132 ].
Acute myeloid leukemia with core binding factor (
CBF-AML ) is characterized by the presence of t(8;21) (q22; q22), or inv(16) (p13q22)/t(16;16), leading to the formation of
RUNX1/RUNX1T1 (AML1/ETO) and
CBFbeta-MYH11 , respectively [
133 ].
ULK1-mediated activation of autophagy can control and delay AML1-ETO9a- driven leukemogenesis in an AML
CASPASE-3 knockout mouse model [
134], suggesting that CASPASE-3 is an important regulator of autophagy in AML. The results of these studies highlight the different roles of autophagy in the initiation, progression and chemotherapy responses in AML cells, depending on the different type of aberrant oncoprotein.
4.2. Gene mutations in AML and autophagy
Among the most common genetic alterations in AML, mutation of the tyrosine kinase 3 ( FLT3 ) gene occurs in approximately 30% of AML cases.
The most frequent aberrations affecting the
FLT3 gene , associated with a poor prognosis in AML, are internal tandem duplication (
FLT3 -ITD) in the juxtamembrane domain and point mutations, involving the tyrosine kinase domain of
FLT3 (
FLT3 -TKD) [
135 ].
Expression of FLT3 -ITD increases basal autophagy in AML cells through a mechanism involving the transcription factor ATF4 (activating transcription factor 4) [
136 ]. Furthermore, the inhibition of autophagy in
FLT3 cells-TKD, which are resistant to the FLT3 inhibitor quizartinib (AC220), also inhibits proliferation both in vitro and in vivo [
136 ]. More recently, the acquired D835Y mutation induced resistance to the FLT3 inhibitor sorafenib and activated autophagy in
FLT3 -ITD-positive cell lines. By inhibiting autophagy, the authors were able to overcome sorafenib resistance in
FLT3 -ITD-positive AML, improving its efficacy [
137 ]. Recently, a study demonstrated that inhibition of autophagy reduces the repopulation potential of mitochondrial storage-associated
FLT3 -ITD AML LSCs [ 138]. Furthermore, the authors demonstrated that autophagy inhibition enhances p53 activity and enhances TKI-mediated inhibition of AML progenitors [
138 ].
Autophagy not only contributes to the downstream proliferation of the FLT3-ITD receptor but may also be involved in the degradation of the mutated receptor. Indeed, in one study, the frequent activation of the RET receptor tyrosine kinase was observed in different subtypes of AML [
139 ]. RET mediates the suppression of autophagy in an mTORC1-dependent manner, leading to the stabilization of the mutant FLT3 receptor. Genetic or pharmacological inhibition of RET reduced the growth of FLT3-dependent AML cells, with upregulation of autophagy and depletion
of FLT3 [
139]. These results suggest that restoration of autophagy in FLT3-dependent AML may result in degradation of mutant FLT3 and thus may represent an attractive therapeutic approach. Inhibition of the FLT3-ITD protein has also been shown to lead to increased ceramide synthesis and mediate ceramide-dependent mitophagy, leading to AML cell death [140
, 141
] .
Mutations in KIT are associated with increased leukemia cell proliferation and an increased risk of AML recurrence [
142 ,
143 ]. A recent study reported that the
KIT D816V mutation in AML cells increases basal autophagy, stimulating AML cell proliferation and survival via STAT3 signaling [
144 ]. A different point mutation in c-
KIT (N822K T > A) constitutively activates this receptor, making AML cells highly sensitive to sunitinib (a tyrosine kinase inhibitor), resulting in AML cell death through the activation of both apoptosis and autophagic processes [
145 ].
Mutations in
NPM1 (nucleophosmin 1) are the most frequent genetic alterations in adult AML, responsible for the aberrant localization of the NPM1 protein in the cytoplasm [
146 ].
The increased autophagic activity found in NPM1 -mutated AML cells is involved in leukemic cell survival [
147 ]. The
NPM1 mutant can also interact with the tumor suppressor protein PML (leukemia pro-myelocytic protein), leading to the delocalization and stabilization of PML which, in turn, can activate autophagy via AKT signaling [147
] . In another study, it was shown that in AML patients carrying the mutant
NPM1 , the glycolytic enzyme PKM2 (pyruvate kinase M2) induced autophagy through phosphorylation of the autophagic protein Beclin 1, contributing to cell survival [
148 ]. Finally, the NPM1 mutant protein can also interact with the autophagic protein ULK1, stimulating TRAF6-dependent ubiquitination of ULK1 via miR-146, thereby maintaining ULK1 stability and functionality and promoting autophagic cell survival [149
] . Furthermore, the expression of RASGRP3, a protein associated with tumor progression, was observed to be upregulated in AML patients with
NPM1 mutation compared to AML patients without mutant
NPM1 . The authors demonstrated that
NPM1 -mut blocks the degradation of RASGRP3 protein through binding to the E3 ubiquitin ligase MID1 protein, leading to overexpression of RASGRP3, as well as promoting downstream activation of EGFR-STAT3, which in turn promotes proliferation and autophagy in AML cells [150
] .
Alterations of the tumor suppressor gene
TP53 are found in approximately 5-15% of AML cases and frequently in older patients [
151 ,
152 ].
It has been proposed that the role of autophagy in the development of AML may be determined by TP53 status . For wild-type AML
TP53 , researchers demonstrated that pharmacological blockade of autophagy achieves therapeutic benefits, whereas AMLs harboring
TP53 mutations fail to respond to autophagy inhibition by hydroxychloroquine (HCQ) [
153 ,
154]. The use of autophagic inhibitors may be a potential therapeutic strategy to use, particularly for the treatment of wild-type
TP53 AML. For AML with
TP53 mutations , autophagic pathways may be a therapeutic option to be used for the elimination of the
TP53 T mutant .
Another study demonstrated that stimulation of macroautophagy by 17-AAG, an HSP90 inhibitor, results in the degradation of TP53 R248Q in AML cells and also enhances the transcription of autophagy-associated genes [155
] . Furthermore, accumulating evidence indicates that TP53 activated by a variety of cellular stresses can trigger autophagy through the transactivation of pro-autophagic genes, including DRAM1
( DNA damage regulated autophagy modulator 1),
SESN1 (sestrin 1), and ( sestrin
SESN2 ) [
155 ,
156 ,
157 ,
158 ].
A recent study highlighted the role of autophagy in AML cells, in the context of p53-mediated apoptosis, which is associated with increased cytotoxicity to treatment with MDM2 inhibitors and Ara-C when miR-10a is inhibited [159
] . The antileukemic strategy based on the use of MDM2/X inhibitors in wild-type p53 tumors to restore the normal and active conformation of p53, MDM2 and MDMX has not been extensively tested [160
] . Therefore, the use of a combination of treatments, including MDM2 inhibitors with autophagic modulators, could be a novel strategy to improve the treatment of wild-type
p53 AML .
Pharmacological treatments that modulate autophagy in AML patients carrying
p53 mutations participate in the degradation of aberrant p53 proteins. Point mutation of
TP53 at the amino acid residue R428 (R248Q), with gain-of-function activity, gives rise to malignant activity in lung cancer cells [
161 ] and a loss of tumor suppressor function in AML [
162 ].
Interestingly, treatment with the Hsp90 inhibitor (17-AAG) results in the activation of chaperone-mediated autophagy, which induces the degradation of the aberrant protein p53R248Q in AML cells. Notably, under conditions of metabolic stress, 17-AAG induces the interaction between p53R248Q and the chaperone protein Hsc70, triggering chaperone-mediated autophagy to degrade p53R248Q [
155 ]. These data open new opportunities for future studies that could elucidate the functional involvement of different types of autophagy and their connection with the molecular mechanisms for improving anticancer therapies against AML harboring the different TP53 variants
.Recent advances in bioinformatics have enabled the identification of several epigenetic mutations affecting AML, including
IDH1/2 , Tet methylcytosine dioxygenase 2 (
TET2 ), DNA methyltransferase 3A (
DNMT3A ) and
ASXL1 , all of which are associated with the pathogenesis of AML [
163 ,
164 ,
165]. IDH proteins are isocitrate dehydrogenases, implicated in various biological processes, such as energy metabolism, histone demethylation, DNA modification, and adaptation to hypoxia. Further studies are needed to investigate innovative therapies based on targeted autophagy in combination with DNA hypomethylation to treat AMLs harboring certain types of epigenetic alterations.
Mutations in the DNMT3A gene , an enzyme involved in CpG dinucleotide methylation, are present in 20-23% of adult patients with de novo AML [
165 ]. Several studies have shown that treatment of AML patients with DNA methyltransferase-inhibiting agents, such as azacitidine (5-aza-2′-deoxycytidine), induces autophagic activity in AML leukemia cells [166
] . A study in a
DNMT3A R878H conditional knock-in mouse model, used to predict the specific long noncoding RNAs (lncRNAs) regulated by
DNMT3A mutations in AML, first identified 23
lncRNAsdifferentially expressed, then downstream target genes regulated by these lncRNAs, including
ATP6V1A , a critical autophagy-related gene, overexpression of which is associated with poor prognosis in AML [
167 ].
However, there is still little evidence for a direct involvement of DNMT3A gene mutations with autophagic activity in AML.
Further studies are needed to understand the functional significance of autophagy associated with different gene mutations in AML cells.
This entry is adapted from the peer-reviewed paper 10.3390/cells12111553