Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Neurosciences
Group I metabotropic glutamate receptors (mGluRI), including mGluR1 and mGluR5 subtypes, modulate essential brain functions by affecting neuronal excitability, intracellular calcium dynamics, protein synthesis, dendritic spine formation, and synaptic transmission and plasticity.
- : mGluR1
- mGluR5
- synaptic plasticity
- glutamatergic transmission
- mGluR1
1. Introduction
Group I metabotropic glutamate receptors (mGluRI), including mGluR1 and mGluR5 subtypes, modulate essential brain functions by affecting neuronal excitability, intracellular calcium dynamics, protein synthesis, dendritic spine formation, and synaptic transmission and plasticity.
2. Group 1 Metabotropic Glutamate Receptors (mGluRI)
mGluR1 and mGluR5 are G protein-coupled receptors (GPCR) constituting the group I metabotropic glutamate receptor (mGluRI). mGluR1 is present in four isoforms (mGluR1α, β, γ, δ) whereas mGluR5 exists in two variants (mGluR5α and mGluR5β). The various isoforms, arising by alternative splicing, mainly differ in the C-terminal intracellular tail [1]. The mGluRI distinction from other mGluR groups (group II, including mGluR2 and mGluR3, and group III, comprising mGluR4, mGluR6, mGluR7, mGluR8) is based on amino-acid homology, agonist binding, and signaling pathways downstream to receptor activation [1].
Canonical mGluRI signaling is driven by the Gq/G11-dependent activation of phospholipase C β (PLCβ), inducing the hydrolysis of phosphoinositides and generation of inositol 1,4,5-trisphosphate (IP3) and diacyl-glycerol (DAG). This pathway leads to the Ca2+ intracellular mobilization from internal stores and activation of protein kinase C (PKC) [1]. In addition, mGluRI can activate several other pathways, and also G protein-independent mechanisms, by means of the interaction with specific adaptor proteins that recruit distinct signaling components [1]. G protein-independent mechanisms mainly lie on β-arrestin binding, favored by the receptor phosphorylation by G-protein-coupled receptor kinases (GRKs). Globally, mGluRI can activate a multifaceted list of effectors, including phospholipase D (PLD), protein kinases pathways such as mitogen-activated protein kinase/extracellular receptor kinase (MAPK/ERK), the mammalian target of rapamycin (mTOR)/p70S6 kinase pathway, casein kinase 1, cyclin-dependent protein kinase 5, and Jun kinase (JUNK) [1,2] (Figure 1).
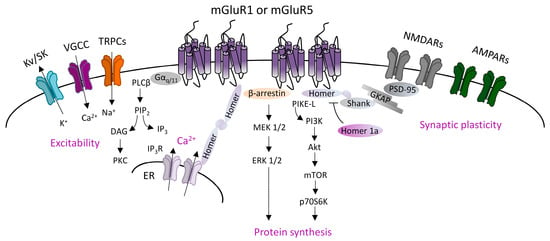
Figure 1. Group 1 metabotropic glutamate receptors (mGluRI) signaling. Scheme of the principal mGluR1 and mGluR5 signaling pathways, showing that Gq/11-dependent activation of phospholipase C β (PLCβ) mediates phosphatidylinositol hydrolysis with the generation of diacylglycerol (DAG) (that activates protein kinase C, PKC) and inositol-1,4,5-trisphosphate (IP3) (that fosters Ca2+ intracellular release from internal stores by acting on IP3R receptors on the endoplasmic reticulum). mGluR1/5, through Gq/11-dependent mechanisms, also modulates ion channels, such as transient receptor potential channels (TRPCs), voltage-gated Ca2+ channels (VGCC), and different types of K+ channels (Kv or SK), thus affecting neuronal excitability. Additional G protein-independent mechanisms involve the recruitment of β-arrestin or other scaffolding/adaptor proteins such as Homer long isoforms, that provide mGluRI coupling with other effectors, thus fostering activation of signaling pathways, such as the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K-Akt-mTOR) kinase pathway, or MEK1/2-ERK 1/2 pathway, both involved in mechanisms promoting protein synthesis.
mGluR1 and mGluR5 are mainly localized in the postsynaptic densities (PSD) in perisynaptic zones [3,4,5], where they form multiprotein complexes by interacting with other membrane proteins and downstream effectors. Different intracellular scaffolding proteins interacting with mGluR1 and mGluR5 have been identified. An important group is constituted by the family of Homer proteins, including long isoforms (Homer 1b, 1c, 2, 3), able to make a large multimeric assembly in the PSD, and the shorter isoform Homer 1a, that does not form protein complexes but antagonizes Homer longer isoforms connections [6,7]. Homer longer isoforms bond mGluR1 or mGluR5 to their principal signaling effectors, such as PLC, PI3K and the PI3K enhancer, PIKE-L, as well as the IP3 receptor, and various classes of ion channels including transient receptor potential-like channels (TRPCs), voltage-gated calcium channels (VGCC), and M-type potassium channels [8,9,10]. Moreover, Homer long isoforms, by linking other scaffolding proteins (most notable are PSD-95 and Shank), directly associate mGluR1/5 to other membrane receptors, including NMDARs. Besides the Homer family, other identified mGluR1- and mGluR5-interacting proteins include tamalin, norbin, preso-1, calmodulin, neuronal calcium-binding protein 2 (NECAB2), calcineurin inhibitor protein (CAIN), the ubiquitin ligase Siah-1A, various protein kinases, including PKC, GRK2, CaMKII, and cytoskeletal components, such as 4.1 G and Filamin-A [8,9,10].
mGluR1 and mGluR5 also bind intracellular regulatory proteins, such as GPCR kinases (GRKs) (GRK2 and GRK3 subtypes for mGluR5, and GRK2, GRK4, and GRK5 for mGluR1) [11,12,13], which control mGluRI internalization by phosphorylation [14], or members of the family of the regulator of G-protein signaling (RGS) (such as RGS-4), which increase the GTPase activity of Gαq and uncouple G protein-linked effectors, and thus switch-off mGluRI signaling [15].
Based on common signaling mechanisms, mGluR1 and mGluR5 have been often regarded as interchangeable, but it is now overt that they can either have separate functions [16,17] or be cross-talking, by acting occasionally in a cooperative [18] or otherwise antagonistic manner [19,20]. mGluR1 and mGluR5 are active as homodimers, i.e., mGluR1-mGluR1 and mGluR5-mGluR5, and additionally the two subtypes can be associated among them in the heterodimer mGluR1-mGluR5, or with other GPCRs belonging to adenosine-, GABA-, and dopamine receptors [21,22,23,24,25,26], by forming dimers such as mGluR1-A1 [23], mGluR1-GABAB [24], and mGluR5-D1 [25], or a trimeric receptor complex, such as mGluR5-A2A-D2 [26]. Furthermore, mGluR1/5 can assembly with ligand-gated ion channels, such as NMDARs [27], and interplay with other ion channels beyond TRPCs, N-type Ca2+ channels, and M-type K+ channels, such as small conductance calcium-activated potassium (SK) channels [28,29], and acid-sensing ion channels 1a (ASIC1a) [30]. Furthermore, mGluR1/5 cross-talk with receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR) and ErbBs, that are receptors for the neurotrophic factors EGF and neuregulins, respectively [31].
mGluR1 and mGluR5 are often co-expressed in the various brain areas and cellular populations, but they can also have different levels or segregate expression in distinct cellular populations [32,33]. Though mGluR1 and mGluR5 are usually described as postsynaptic, they can also be presynaptic [33], and are expressed in astrocytes and microglial cells in several brain areas in physiological and pathological conditions [34], thus implying a more complex scenario of potential mGluR1/5-engaged mechanisms. Furthermore, in addition to plasma membrane-anchored receptors, functional mGluR1/5 pools are located in intracellular compartments, in the endoplasmic reticulum (ER), nuclear, and mitochondrial membranes, wherein they can activate different signaling pathways, thus supporting the cytoplasm-to-nuclear translocation or mGluR1/5 mitochondrial functions [35,36,37].
In conclusion, mGluR1- and mGluR5 interplay with other membrane receptors, ion channels, intracellular scaffolding proteins, regulatory proteins, and signaling pathways, as well as differences in their cellular and sub-cellular localization rule definite roles in different contexts, shaping mGluR1/5 impact on brain functions.
This entry is adapted from the peer-reviewed paper 10.3390/cells12121588
This entry is offline, you can click here to edit this entry!
