Twice-a-year, large-scale movement of billions of birds across latitudinal gradients is one of the most fascinating behavioral phenomena seen among animals. These seasonal voyages in autumn southwards and in spring northwards occur within a discrete time window and, as part of an overall annual itinerary, involve close interaction of the endogenous rhythm at several levels with prevailing photoperiod and temperature. The overall success of seasonal migrations thus depends on their close coupling with the other annual sub-cycles, namely those of the breeding, post-breeding recovery, molt and non-migratory periods. There are striking alterations in the daily behavior and physiology with the onset and end of the migratory period, as shown by the phase inversions in behavioral (a diurnal passerine bird becomes nocturnal and flies at night) and neural activities. Interestingly, there are also differences in the behavior, physiology and regulatory strategies between autumn and spring (vernal) migrations. Concurrent molecular changes occur in regulatory (brain) and metabolic (liver, flight muscle) tissues, as shown in the expression of genes particularly associated with 24 h timekeeping, fat accumulation and the overall metabolism.
- bird
- brain
- gene expression
- heritability
- migration
- seasonal
1. Introduction
2. Migration: A Heritable Seasonal Behavior
2.1. An Innate Migratory Template
2.2. Heritable Migratory Activity Pattern
2.3. Heritable Arrival Dates
3. Unraveling the Genetic Control of Migratory Behavior
3.1. Gene Polymorphism
3.2. Gene Transcription
3.3. Global Gene Analyses
This entry is adapted from the peer-reviewed paper 10.3390/genes14061191
References
- Lack, D. Bird migration and natural selection. Oikos 1968, 19, 1–9.
- Salomonsen, F. The evolutionary significance of bird migration. Dan. Biol. Medd. 1955, 22, 1–62.
- Alerstam, T. Bird Migration; Cambridge University Press: Cambridge, UK, 1991.
- Berthold, P. Control of Bird Migration; Chapman and Hall: London, UK, 1996.
- Pulido, F.; Berthold, P. Quantitative genetic analysis of migratory behavior. In Avian Migration; Berthold, P., Gwinner, E., Sonnenschein, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 53–77.
- Harris, M.P. Abnormal migration and hybridization of Larus argentatus and L. fuscus after fostering experiments. Ibis 1969, 112, 488–498.
- Berthold, P.; Wiltschko, W.; Miltenberger, H.; Querner, U. Genetic transmission of migratory behavior into a nonmigratory bird population. Experientia 1990, 46, 107–108.
- Liedvogel, M.; Akesson, S.; Bensch, S. The genetics of migration on the move. Trends Ecol. Evol. 2011, 26, 561–569.
- Valikangas, I. Finnische Zugvogel aus englischen Vogeleiern. Vogelzug 1933, 4, 159–166.
- Nice, M.M. Zur Naturgeschichte des Singammers. J. Ornithol. 1934, 82, 1–96.
- Nice, M.M. Studies in the life history of the song sparrow. Vol I A population study of the song sparrow. Trans. Linn. Soc. N. Y. 1937, 4–6, 1–247.
- Berthold, P. Genetic control of migratory behavior in birds. Trends Ecol. Evol. 1991, 6, 254–257.
- Berthold, P.; Querner, U. Genetic basis of migratory behavior in European warblers. Science 1981, 212, 77–79.
- Berthold, P.; Helbig, A.J.; Mohr, G.; Querner, U. Rapid microevolution of migratory behaviour in a wild bird species. Nature 1992, 360, 668–670.
- Vega, M.L.; Willemoes, M.; Thomson, R.L.; Tolvanen, J.; Rutila, J.; Samaš, P.; Strandberg, R.; Grim, T.; Fossøy, F.; Stokke, B.G.; et al. First-time migration in juvenile common cuckoos documented by satellite tracking. PLoS ONE 2016, 11, e0168940.
- Berthold, P.; Helbig, A.J. The genetics of bird migration: Stimulus, timing, and direction. Ibis 1992, 134 (Suppl. S1), 35–40.
- Berthold, P.; Pulido, F. Heritability of migratory activity in a natural bird population. Proc. R. Soc. Lond. B 1994, 257, 311–315.
- Berthold, P. Endogenous component of annual cycles of migration and moult. In Acta XVIII Congressus Internationalis Ornithologici; Ilyichev, V.D., Gavrilov, V.M., Eds.; Nauka: Moscow, Ruassia, 1985; pp. 922–929.
- Gwinner, E. Circadian and circannual programmes in avian migration. J. Exp. Biol. 1996, 199, 39–48.
- Berthold, P.; Gwinner, E.; Klein, H.; Westrich, P. Beziehungen zwischen Zugunruhe und Zugablauf bei Garten und Monchsgrasmucke (Sylvia borin und S. atricapilla). Z. Tierpsychol. 1972, 30, 26–35.
- Hedenström, A. Migration routes and wintering areas of willow warblers Phylloscopus trochilus (L.) ringed in Fennoscandia. Ornis Fenn. 1987, 64, 137.
- Lerche-Jørgensen, M.; Willemoes, M.; Tøttrup, A.P.; Snell, K.R.; Thorup, K. No apparent gain from continuing migration for more than 3000 kilometres: Willow warblers breeding in Denmark winter across the entire northern Savannah as revealed by geolocators. Mov. Ecol. 2017, 5, 17.
- Zhao, T.; Ilieva, M.; Larson, K.; Lundberg, M.; Neto, J.M.; Sokolovskis, K.; Åkesson, S.; Bensch, S. Autumn migration direction of juvenile willow warblers (Phylloscopus t. trochilus and P. t. acredula) and their hybrids assessed by qPCR SNP genotyping. Mov. Ecol. 2020, 8, 22.
- Sokolovskis, K.; Lundberg, M.; Åkesson, S.; Willemoes, M.; Zhao, T.; Caballero-Lopez, V.; Bensch, S. Migration direction in a songbird explained by two loci. Nat. Comm. 2023, 14, 165.
- Morrison, C.A.; Alves, J.A.; Gunnarsson, T.G.; Porisson, B.; Gill, J.A. Why do earlier arriving migratory birds have better breeding success? Ecol. Evol. 2019, 9, 8856–8864.
- Aebischer, A.; Perrin, N.; Krieg, M.; Studer, J.; Meyer, D.R. The role of territory choice, mate choice and arrival date on breeding success in the Savi’s Warbler, Locustella luscinioides. J. Avian Biol. 1996, 27, 143–152.
- Currie, D.; Thompson, D.; Burke, T. Patterns of territory settlement and consequences for breeding success in the northern wheatear Oenanthe oenanthe. Ibis 2000, 142, 389–398.
- McKellar, A.E.; Marra, P.P.; Ratcliffe, L.M. Starting over: Experimental effects of breeding delay on reproductive success in early-arriving male American redstarts. J. Avian Biol. 2013, 44, 495–503.
- Norris, D.R.; Marra, P.P.; Kyser, T.K.; Sherry, T.W.; Ratcliffe, L.M. Tropical winter habitat limits reproductive success on the temperate breeding grounds in a migratory bird. Proc. Royal Soc. B Biol. Sci. 2004, 271, 59–64.
- Rockwell, S.M.; Bocetti, C.I.; Marra, P.P. Carry-over effects of winter climate on spring arrival date and reproductive success in an endangered migratory bird, Kirtland’s Warbler (Setophaga kirtlandii). Auk 2012, 129, 744–752.
- Velmala, W.; Helle, S.; Ahola, M.P.; Klaassen, M.; Lehikoinen, E.; Rainio, K.; Sirkia, P.M.; Laaksonen, T. Natural selection for earlier male arrival to breeding grounds through direct and indirect effects in a migratory songbird. Ecol. Evol. 2015, 5, 1205–1213.
- Tarka, M.; Hansson, B.; Hasselquist, D. Selection and evolutionary potential of spring arrival phenology in males and females of a migratory songbird. J. Evol. Biol. 2015, 28, 1024–1038.
- Bowers, E.K.; Grindstaff, J.L.; Soukup, S.S.; Drilling, N.E.; Eckerle, K.P.; Sakaluk, S.K.; Thompson, C.F. Spring temperatures influence selection on breeding date and the potential for phenological mismatch in a migratory bird. Ecology 2016, 97, 2880–2891.
- Visser, M.E.; Gienapp, P.; Husby, A.; Morrisey, M.; de la Hera, I.; Pulido, F.; Both, C. Effects of spring temperatures on the strength of selection on timing of reproduction in a long-distance migratory bird. PLoS Biol. 2015, 13, e1002120.
- Kullberg, C.; Fransson, T.; Hedlund, J.; Jonzén, N.; Langvall, O.; Nilsson, J.; Bolmgren, K. Change in spring arrival of migratory birds under an era of climate change, Swedish data from the last 140 years. Ambio 2015, 44 (Suppl. S1), S69–S77.
- Mueller, J.C.; Pulido, F.; Kempenaers, B. Identification of a gene associated with avian migratory behavior. Proc. R. Soc. B 2011, 278, 2848–2856.
- Bazzi, G.; Ambrosini, R.; Caprioli, M.; Costanzo, A.; Liechti, F.; Gatti, E.; Gianfranceschi, L.; Podofillini, S.; Romano, A.; Romano, M.; et al. Clock gene polymorphism and scheduling of migration: A geolocator study of the barn swallow Hirundo rustica. Sci. Rep. 2015, 5, 12443.
- Saino, N.; Bazzi, G.; Gatti, E.; Caprioli, M.; Cecere, J.G.; Possenti, C.D.; Galimberti, A.; Orioli, V.; Bani, L.; Rubolini, D.; et al. Polymorphism at the Clock gene predicts phenology of long-distance migration in birds. Mol. Ecol. 2015, 24, 1758–1773.
- Peterson, M.P.; Abolins-Abols, M.; Atwell, J.W.; Rice, R.J.; Milá, B.; Ketterson, E.D. Variation in candidate genes CLOCK and ADCYPA1 does not consistently predict differences in migratory behaviour in the songbird genus Junco. F1000 Res. 2013, 2, 115.
- Parody-Merino, Á.M.; Battley, P.F.; Conklin, J.R.; Fidler, A.E. No evidence for an association between Clock gene allelic variation and migration timing in a long-distance migratory shorebird (Limosa lapponica baueri). Oecologia 2019, 191, 843–859.
- Kramer, G.R.; Streby, H.M.; Peterson, S.M.; Lehman, J.A.; Buehler, D.A.; Wood, P.B.; McNeil, D.J.; Larkin, J.L.; Andersen, D.E. Nonbreeding isolation and population-specific migration patterns among three populations of Golden-winged Warblers. Condor Ornithol. Appl. 2017, 119, 108–121.
- Kramer, G.R.; Andersen, D.E.; Buehler, D.A.; Wood, P.B.; Peterson, S.M.; Lehman, J.A.; Aldinger, K.R.; Bulluck, L.P.; Harding, S.; Jones, J.A.; et al. Population trends in Vermivora warblers are linked to strong migratory connectivity. Proc. Natl. Acad. Sci. USA 2018, 115, E3192–E3200.
- Toews, D.P.L.; Taylor, S.A.; Streby, H.M.; Kramer, G.R.; Lovette, I.J. Selection on VPS13A linked to migration in a songbird. Proc. Nat. Acad. Sci. USA 2019, 116, 18272–18274.
- Gu, Z.; Pan, S.; Lin, Z.; Hu, L.; Dai, X.; Chang, J.; Xue, Y.; Su, H.; Long, J.; Sun, M.; et al. Climate-driven flyway changes and memory-based long-distance migration. Nature 2021, 591, 259–264.
- Merlin, C.; Liedvogel, M. The genetics and epigenetics of animal migration and orientation: Birds, butterflies and beyond. J. Exp. Biol. 2019, 222 (Suppl. S1), jeb191890.
- Delmore, K.; Illera, J.C.; Pérez-Tris, J.; Segelbacher, G.; Ramos, J.S.L.; Durieux, G.; Ishigohoka, J.; Liedvogel, M. The evolutionary history and genomics of European blackcap migration. eLife 2020, 9, e54462.
- Bossu, C.M.; Heath, J.A.; Kaltenecker, G.S.; Helm, B.; Ruegg, K.C. Clock linked genes underlie seasonal migratory timing in a diurnal raptor. Proc. R. Soc. B 2022, 289, 20212507.
- Newton, I. The Migration Ecology of Birds; Academic Press: London, UK, 2007.
- Tryjanowski, P.; Yosef, R. Differences between the spring and autumn migration of the red-backed shrike Lanius collurio: Record from the Eilat Stopover (Israel). Acta Ornithol. 2002, 37, 85–90.
- Nilsson, C.; Klaassen, R.H.G.; Alerstam, T. Differences in speed and duration of bird migration between spring and autumn. Am. Nat. 2013, 181, 837–845.
- Cornelius, J.M.; Boswell, T.; Eiermann, S.J.; Breuner, C.W.; Ramenofsky, M. Contribution of endocrinology to the migration life history of birds. Gen. Comp. Endocrinol. 2013, 190, 47–60.
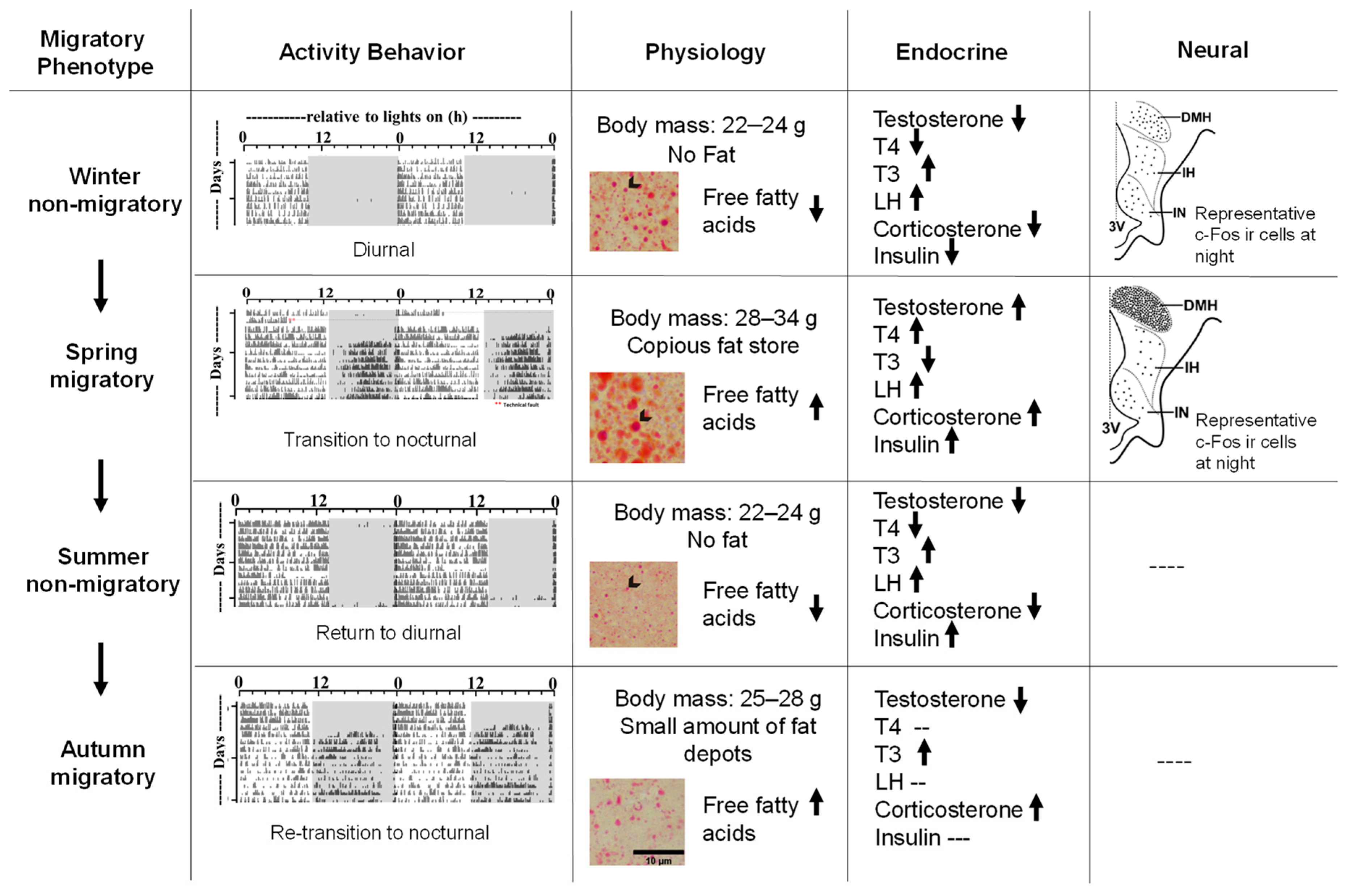
- Sharma, A.; Singh, D.; Malik, S.; Gupta, N.J.; Rani, S.; Kumar, V. Difference in control between spring and autumn migration in birds: Insight from seasonal changes in hypothalamic gene expression in captive buntings. Proc. R. Soc. B 2018, 285, 1531.
- Sharma, A.; Kumar, V. Metabolic plasticity mediates differential responses to spring and autumn migrations: Evidence from gene expression patterns in migratory buntings. Exp. Physiol. 2019, 104, 1841–1857.
- Rastogi, A.; Kumari, Y.; Rani, S.; Kumar, V. Neural correlates of migration: Activation of hypothalamic clock(s) in and out of migratory state in the blackheaded bunting (Emberiza melanocephala). PLoS ONE 2013, 8, e70065.
- Jain, N.; Kumar, V. Changes in food intake, body weight, gonads and plasma concentrations of thyroxine, luteinizing hormone and testosterone in captive male buntings exposed to natural daylengths at 29° N. J. Biosci. 1995, 20, 417–426.
- Mishra, I.; Singh, D.; Kumar, V. Daily levels and rhythm in circulating corticosterone and insulin are altered with photostimulated seasonal states in night-migratory blackheaded buntings. Horm. Behav. 2018, 94, 114–123.
- Lundberg, M.; Boss, J.; Canbäck, B.; Liedvogel, M.; Larson, K.W.; Grahn, M.; Akesson, S.; Bensch, S.; Wright, A. Characterisation of a transcriptome to find sequence differences between two differentially migrating subspecies of the willow warbler Phylloscopus trochilus. BMC Genom. 2013, 14, 330.
- Fudickar, A.M.; Peterson, M.P.; Greives, T.J.; Atwell, J.W.; Bridge, E.S.; Ketterson, E.D. Differential gene expression in seasonal sympatry: Mechanisms involved in diverging life histories. Biol. Lett. 2016, 12, 20160069.
- Boss, J.; Liedvogel, M.; Lundberg, M.; Olsson, P.; Reischke, N.; Naurin, S.; Akesson, S.; Hasselquist, D.; Wright, A.; Grahn, M.; et al. Gene expression in the brain of a migratory songbird during breeding and migration. Mov. Ecol. 2016, 4, 4.
- Johnston, R.A.; Paxton, K.L.; Moore, F.R.; Wayne, R.K.; Smith, T.B. Seasonal gene expression in a migratory songbird. Mol. Ecol. 2016, 25, 5680–5691.
- Franchini, P.; Irisarri, I.; Fudickar, A.; Schmidt, A.; Meyer, A.; Wikelski, M.; Partecke, J. Animal tracking meets migration genomics: Transcriptomic analysis of a partially migratory bird species. Mol. Ecol. 2017, 26, 3204–3216.
