Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Endoscopic Ultrasound (EUS) is widely used for the diagnosis of bilio-pancreatic and gastrointestinal (GI) tract diseases, for the evaluation of subepithelial lesions, and for sampling of lymph nodes and solid masses located next to the GI tract. The role of Artificial Intelligence in healthcare in growing.
- endoscopic ultrasound
- artificial intelligence
- biopsy
- pathological diagnosis
1. Introduction
Endoscopic ultrasound (EUS) has revolutionized the field of gastrointestinal (GI) endoscopy by providing high-resolution imaging of the gastrointestinal tract and adjacent anatomical structures. EUS has been widely used for the diagnosis of bilio-pancreatic diseases, staging of GI tract tumors, evaluation of subepithelial lesions, and sampling of lymph nodes and solid masses [1]. EUS-guided fine-needle aspiration (FNA) and biopsy (FNB) have enabled the diagnosis of various malignancies and have greatly improved patient outcomes [2].
However, the accuracy of EUS-guided FNA and FNB largely depends on the skills and experience of the endoscopist and of the pathologist. In recent years, artificial intelligence (AI) has emerged as a promising tool for improving the accuracy and efficiency of EUS-guided tissue sampling and pathological diagnosis [3].
AI refers to the use of computer algorithms to analyze large amounts of data and identify patterns or make predictions. In healthcare, AI has been applied to various tasks, including image recognition, natural language processing, and clinical decision-making [4]. AI algorithms can analyze EUS images and assist with the interpretation of findings, as well as predict the pathological diagnosis of tissue samples obtained by EUS-guided FNA and FNB.
2. AI Algorithms and Image Acquisition
The use of artificial intelligence in EUS image interpretation has shown great potential for improving the accuracy and efficiency of the diagnostic process. AI algorithms can be divided into two main categories: deep learning techniques and machine learning techniques.
Deep learning techniques involve the use of neural networks to learn and recognize patterns in EUS images (Figure 1). These networks are composed of multiple layers of interconnected nodes that allow for the processing of large amounts of data. Convolutional neural networks (CNNs) are a commonly used deep learning technique for image analysis in healthcare [5]. They are designed to identify and extract important features from EUS images and use them to classify or segment the images.
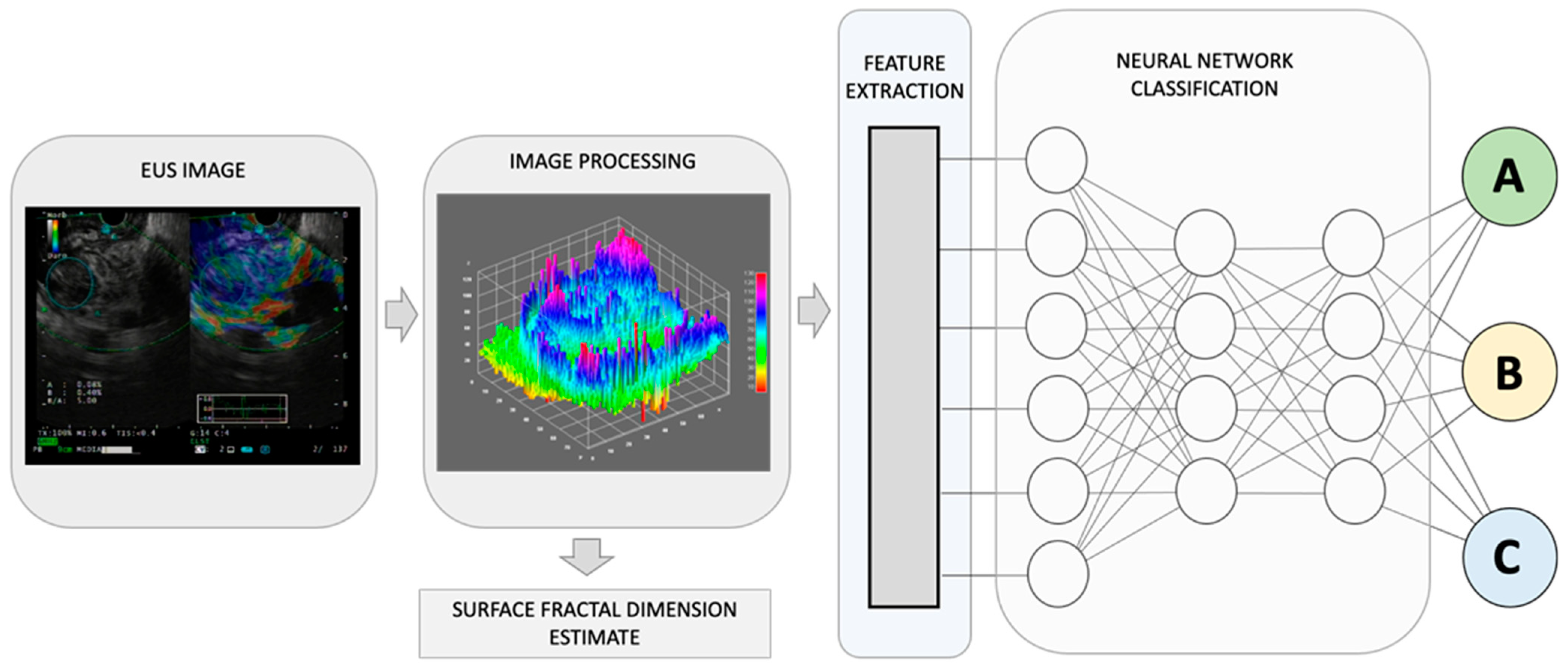
Figure 1. By combining recognized EUS-image features for pancreatic lesion diagnosis with measurements of non-Euclidean anatomical features, significant progress can be made in distinguishing diverse sub-types that have varying outcomes, A, B, and C. The utilization of fractal geometry, specifically the surface fractal dimension, as a measure of the space-filling property of an irregularly shaped structure, can be effectively merged as a feature within an AI-based neuronal network classification system, to achieve a more precise anatomical classifier system.
Machine learning techniques, on the other hand, involve the use of algorithms that can learn from data and make predictions based on that learning. These techniques can be supervised, unsupervised, or semi-supervised. Supervised learning involves the use of labelled data to train an algorithm to recognize patterns in EUS images. Unsupervised learning involves the use of unlabeled data to discover patterns and relationships in the data. Semi-supervised learning combines both supervised and unsupervised learning [6]. In addition to image interpretation, AI can also be used to enhance the acquisition of EUS images. This includes automatic segmentation and image quality improvement.
Automatic segmentation involves the use of AI algorithms to identify and separate different structures in EUS images [7]. This can help to improve the accuracy and efficiency of EUS-guided procedures by providing better visualization of the target area. For example, AI algorithms can be used to automatically segment the pancreas or the lymph nodes in EUS images, allowing for more precise targeting during EUS-guided biopsy [8].
Image quality improvement involves the use of AI algorithms to enhance the clarity and resolution of EUS images. This can help to improve the accuracy of image interpretation and diagnosis. For example, AI algorithms can be used to reduce noise, improve contrast, and sharpen edges in EUS images. AI-enhanced image quality can also help to reduce the variability in image quality between different endoscopists and ultrasound machines, improving the consistency of diagnosis and treatment [9].
3. Lesion Detection and Characterization
3.1. Tumor Identification
Tumor identification is a crucial step in the diagnosis and staging of GI neoplasia, especially in case of pancreatic cancers that may be isoechoic with the surrounding parenchyma or may be hidden by signs of chronic pancreatitis. AI algorithms can assist with tumor identification by analyzing EUS images and identifying suspicious areas that may require biopsy sampling and microscopy observation or further clinical evaluation [10]. Deep learning techniques, such as CNNs, have shown great potential for tumor identification by extracting important features from EUS images and using them to classify or segment the images [11]. Supervised machine learning techniques can also be used to train AI algorithms to recognize specific tumor features, such as shape, size, and vascularity [12][13]. A recent meta-analysis of 10 studies, which involved 1871 patients, evaluated the diagnostic accuracy of AI applied to EUS in detecting pancreatic cancer. The results showed that AI had a high diagnostic sensitivity of 0.92 and specificity of 0.9, with an area under the summary receiver operating characteristics (SROC) curve of 0.95 and a diagnostic odds ratio of 128.9 [14]. These findings suggest that AI-assisted EUS could become an essential tool for the computer-aided diagnosis of pancreatic cancer. However, a relatively small number of studies and enrolled patients make generalizing difficult, and further research is needed to validate these results on a larger scale.
It is known that there are several advantages of incorporating new features in classifying pathological changes using AI. Among these are the following: (a) increased accuracy: the addition of new features to AI models can increase their accuracy in diagnosing and classifying pathological changes; (b) faster diagnosis: AI models with new features can analyze large amounts of data quickly and accurately, allowing for early diagnosis and better patient outcomes with reduced healthcare costs; (c) personalized treatment: AI models with new features can identify subtle differences in disease presentation that may be missed by human eyes. This can lead to more personalized treatment plans that are tailored to the specific needs of each patient; (d) improved decision-making: AI models with new features can provide clinicians with more information and insights into disease pathology, allowing them to make more informed decisions about patient care; and (e) scalability: AI models can be easily scaled to analyze large amounts of data, making them ideal for analyzing large datasets or monitoring patient health over time. This can lead to improved population health management and disease surveillance.
Diseases of various origins, such as inflammatory disorders, tumors, and functional diseases, can result in changes in the structural complexity and dynamic activity patterns. One way to quantify this structural complexity is by measuring the fractal dimension, among other parameters.
The human body is composed of intricate systems and networks, including its most complex structures. It is now widely accepted that the architecture of anatomical entities and their activities exhibit non-Euclidean properties. Natural fractals, including those found in anatomy, possess four distinct characteristics: (a) irregular shape, (b) statistical self-similarity, (c) non-integer or fractal dimension, and (d) scaling properties that depend on the scale of measurement. As anatomical structures do not conform to regular Euclidean shapes, their dimensions are expressed as non-integer values between two integer topological dimensions [15]. Fractal geometry has been shown to be useful in evaluating the geometric complexity of anatomic and imaging patterns observed in both benign and malignant masses (Figure 1).
Recently, Carrara et al. have introduced a new estimator, called the surface fractal dimension, to evaluate the complexity of EUS-Elastography images in differentiating solid pancreatic lesions [16]. The study showed that the surface fractal dimension can distinguish malignant tumors from NETs, unaffected tissues surrounding malignant tumors from NETs, and NETs from inflammatory lesions. This study highlights the importance of incorporating fractal analysis into AI algorithms for the diagnosis and categorization of the diverse array of pancreatic lesions.
3.2. Subepithelial Lesion Evaluation
Subepithelial lesions (SELs) are a common indication to perform EUS, and their diagnosis can be challenging. AI algorithms can assist with SELs evaluation by analyzing EUS images and identifying suspicious lesions that may require biopsy. Deep learning techniques, such as CNNs, have shown great potential for SELs evaluation by extracting important features from EUS images and using them to classify or segment the images. The study by Hirai et al. suggests that an AI system has higher diagnostic performance than experts in differentiating SELs on EUS images [17]. The AI system’s accuracy for classifying five different types of SELs was 86.1%, which was significantly better than that of all endoscopists. In particular, the sensitivity and accuracy of the AI system for detecting gastrointestinal stromal tumors (GISTs) were higher than those of all endoscopists. These findings suggest that AI technology can be a valuable tool to assist in the diagnosis of SELs on EUS images, and may help improve clinical decision-making [17].
3.3. Diagnostic Accuracy
The diagnostic accuracy of AI algorithms may be affected by several factors, such as the quality of the EUS images, the size and location of the lesion, and the expertise of the endoscopists and the pathologists. The results of previous studies have been controversial. In recent meta-analysis, Xiao et al. identified seven studies to assess the diagnostic accuracy of AI-based EUS in distinguishing GISTs from other SELs [18]. The combined sensitivity and specificity of AI-based EUS were 0.93 and 0.78, respectively, with an overall diagnostic odds ratio of 36.74 and an area under the summary receiver operating characteristic curve (AUROC) of 0.94. These results suggest that AI-based EUS showed high diagnostic ability in differentiating GISTs from other SELs and could potentially set a premise for adapting diagnostic capabilities of other disease under EUS.
3.4. Clinical Impact and Limitations
The clinical impact of AI algorithms for lesion detection and characterization in EUS-guided pathological diagnosis is still under investigation. However, studies have reported that AI algorithms can improve the accuracy and efficiency of EUS-guided procedures, reduce the need for unnecessary biopsies, and assist with treatment planning [19]. AI algorithms can also help to reduce the inter-observer variability in lesion detection and characterization between different endoscopists [20]. However, the implementation of AI algorithms in clinical practice may be limited by several factors, such as the availability and cost of AI software, the need for specialized training, concerns about data privacy and security, and most importantly the need for larger studies to establish the accuracy of such systems.
4. Digital Histopathological Diagnosis
The advancement of digital pathology has revolutionized the field of pathology by enabling the acquisition, management, and interpretation of pathological information in a digital format. This transition has been fueled by advances in whole slide imaging (WSI) technology, which allows for the digitization of glass slides at high resolution [21]. The adoption of digital pathology offers numerous advantages, such as improved efficiency, reduced turnaround times, remote consultation, and easy access to archived cases [21]. The ability to store pictures from tissue acquisition, as it happens for radiological imaging, puts the basis to share and use a lot of knowledge from pathological anatomy. Moreover, it sets the stage for the application of AI algorithms to facilitate and enhance diagnostic accuracy in the field of EUS.
WSI involves the scanning of entire histological glass slides to create high-resolution digital images. These digital images can be zoomed in or out and navigated as easily as a glass slide under a microscope. WSI technology has been instrumental in overcoming the challenges of data management in digital pathology, as it allows for efficient storage, retrieval, and sharing of massive amounts of image data [21]. Furthermore, WSI facilitates the standardization of image quality and provides an ideal platform for the application of AI algorithms to analyze the digital images, thereby supporting the development of novel diagnostic tools in endoscopic ultrasound.
In the context of histopathological image analysis, CNNs can be trained to automatically detect and classify the multifarious tissue structures, cellular patterns, and pathological alterations. CNNs are composed of multiple layers of interconnected “neurons”, including convolutional, pooling, and fully connected layers. The hierarchical structure of CNNs allows them to learn complex, high-level features from raw image data, thereby making them particularly suitable for the analysis of intricate histopathological images in endoscopic ultrasound [22][23][24].
In addition to CNNs, other machine learning techniques have been employed in histopathological image analysis for endoscopic ultrasound. These include support vector machines (SVM), random forests, and decision trees, among others. These algorithms can be used to extract and analyze peculiar features from histopathological images, such as texture, shape, and color. By leveraging the strengths of multiple machine learning techniques, ensemble models can be created to improve overall performance and address potential limitations of individual algorithms [25].
The integration of AI algorithms, particularly CNNs, into the field of EUS has the potential to revolutionize the diagnosis and management of various GI tract disorders. As research progresses and these techniques become more refined, AI-based tools are expected to play an increasingly prominent role in the field of EUS and digital pathology.
5. AI in Pathological Diagnosis
5.1. AI Applications in Anatomical Pathology
Tumor grading and staging are critical steps in the management of GI malignancies. Accurate tumor grading and staging are necessary for determining the appropriate treatment plan and predicting patient outcomes. AI algorithms can aid in tumor grading and staging by analyzing EUS images and identifying features that correspond to different tumor stages and grades (Figure 1). Deep learning techniques, such as CNNs, can identify subtle differences in tissue structure and morphology that may not be apparent to the human eye. For example, CNNs can potentially analyze EUS images of pancreatic cancer and differentiate between early-stage and advanced-stage tumors based on changes in tissue texture and vascularity [26][27][28]. AI algorithms can also predict the presence of lymph node metastasis by analyzing EUS images and identifying characteristic features, such as size, shape, and echogenicity. In a study by Săftoiu et al., contrast-enhanced harmonic EUS (CEH-EUS) with time-intensity curve (TIC) analysis and artificial neural network (ANN) processing were used to differentiate pancreatic carcinoma (PC) and chronic pancreatitis (CP) cases [29]. Parameters obtained through TIC analysis were able to differentiate between PC and CP cases and showed good diagnostic results in an automated computer-aided diagnostic system.
Prognostic and predictive biomarker analysis is essential for predicting patient outcomes and determining the most appropriate treatment plan. Prognostic biomarkers are associated with patient outcomes, such as survival or recurrence, while predictive biomarkers are associated with response to specific therapies [30]. Kurita et al. investigated the diagnostic ability of carcinoembryonic antigen (CEA), cytology, and AI using cyst fluid in differentiating malignant from benign pancreatic cystic lesions [31]. AI using deep learning showed higher sensitivity and accuracy in differentiating malignant from benign pancreatic cystic lesions than CEA and cytology.
5.2. Integrating AI in EUS-Guided Tissue Acquisition
EUS-guided fine needle aspiration (EUS-FNA) and EUS-guided fine needle biopsy (EUS-FNB) are commonly used techniques for obtaining tissue samples for pathological diagnosis. The accuracy of EUS-guided tissue acquisition largely depends on the skills and experience of the operator and the quality and size of the tissue samples obtained can vary. The integration of AI in EUS-guided tissue acquisition has the potential to improve the accuracy and efficiency of the procedure, leading to better patient outcomes.
In a study by Inoue et al., an automatic visual inspection method based on supervised machine learning was proposed to assist rapid on-site evaluation (ROSE) for endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) biopsy. The proposed method was effective in assisting on-site visual inspection of cellular tissue in ROSE for EUS-FNA, indicating highly probable areas including tumor cells [32].
Hashimoto et al. evaluated the diagnostic performance of their computer-aided diagnosis system using deep learning in EUS-FNA cytology of pancreatic ductal adenocarcinoma. The deep learning system showed promising results in improving diagnostic performance by step-by-step learning, with higher training volume and more efficient system development required for optimal CAD performance in ROSE of EUS-FNA cytology [33].
Ishikawa et al. developed a new AI-based method for evaluating EUS-FNB specimens in pancreatic diseases using deep learning and contrastive learning. The AI-based evaluation method using contrastive learning was comparable to macroscopic on-site evaluation (MOSE) performed by EUS experts and can be a novel objective evaluation method for EUS-FNB [19].
AI algorithms can potentially assist in EUS-FNA and EUS-FNB by providing real-time feedback to the endoscopist during the procedure. AI algorithms can analyze EUS images in real-time and provide guidance on the optimal location and depth of the needle insertion, as well as feedback on the quality of the tissue sample obtained. AI algorithms can also assist in the selection of the appropriate needle size and type based on the characteristics of the target lesion, such as diameter and location. The expectations of the limit of how AI can help improve variability in an endoscopic procedure can be the forefront need to find applicability from these systems to aid the outcome of a procedure [3].
This entry is adapted from the peer-reviewed paper 10.3390/jcm12113757
References
- Friedberg, S.R.; Lachter, J. Endoscopic ultrasound: Current roles and future directions. World J. Gastrointest. Endosc. 2017, 9, 499–505.
- Sooklal, S.; Chahal, P. Endoscopic Ultrasound. Surg. Clin. N. Am. 2020, 100, 1133–1150.
- Liu, E.; Bhutani, M.S.; Sun, S. Artificial intelligence: The new wave of innovation in EUS. Endosc. Ultrasound 2021, 10, 79–83.
- Yu, K.-H.; Beam, A.L.; Kohane, I.S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018, 2, 719–731.
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629.
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515.
- Iwasa, Y.; Iwashita, T.; Takeuchi, Y.; Ichikawa, H.; Mita, N.; Uemura, S.; Shimizu, M.; Kuo, Y.-T.; Wang, H.-P.; Hara, T. Automatic Segmentation of Pancreatic Tumors Using Deep Learning on a Video Image of Contrast-Enhanced Endoscopic Ultrasound. J. Clin. Med. 2021, 10, 3589.
- Sinkala, M.; Mulder, N.; Martin, D. Machine Learning and Network Analyses Reveal Disease Subtypes of Pancreatic Cancer and their Molecular Characteristics. Sci. Rep. 2020, 10, 1212.
- Simsek, C.; Lee, L.S. Machine learning in endoscopic ultrasonography and the pancreas: The new frontier? Artif. Intell. Gastroenterol. 2022, 3, 54–65.
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157.
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444.
- Murali, N.; Kucukkaya, A.; Petukhova, A.; Onofrey, J.; Chapiro, J. Supervised Machine Learning in Oncology: A Clinician’s Guide. Dig. Dis. Interv. 2020, 4, 73–81.
- Shao, D.; Dai, Y.; Li, N.; Cao, X.; Zhao, W.; Cheng, L.; Rong, Z.; Huang, L.; Wang, Y.; Zhao, J. Artificial intelligence in clinical research of cancers. Brief. Bioinform. 2022, 23, bbab523.
- Dumitrescu, E.A.; Ungureanu, B.S.; Cazacu, I.M.; Florescu, L.M.; Streba, L.; Croitoru, V.M.; Sur, D.; Croitoru, A.; Turcu-Stiolica, A.; Lungulescu, C.V. Diagnostic Value of Artificial Intelligence-Assisted Endoscopic Ultrasound for Pancreatic Cancer: A Systematic Review and Meta-Analysis. Diagn. Basel Switz. 2022, 12, 309.
- Glenny, R.W.; Robertson, H.T.; Yamashiro, S.; Bassingthwaighte, J.B. Applications of fractal analysis to physiology. J. Appl. Physiol. 1991, 70, 2351–2367.
- Carrara, S.; Di Leo, M.; Grizzi, F.; Correale, L.; Rahal, D.; Anderloni, A.; Auriemma, F.; Fugazza, A.; Preatoni, P.; Maselli, R.; et al. EUS elastography (strain ratio) and fractal-based quantitative analysis for the diagnosis of solid pancreatic lesions. Gastrointest. Endosc. 2018, 87, 1464–1473.
- Hirai, K.; Kuwahara, T.; Furukawa, K.; Kakushima, N.; Furune, S.; Yamamoto, H.; Marukawa, T.; Asai, H.; Matsui, K.; Sasaki, Y.; et al. Artificial intelligence-based diagnosis of upper gastrointestinal subepithelial lesions on endoscopic ultrasonography images. Gastric Cancer 2022, 25, 382–391.
- Ye, X.H.; Zhao, L.L.; Wang, L. Diagnostic accuracy of endoscopic ultrasound with artificial intelligence for gastrointestinal stromal tumors: A meta-analysis. J. Dig. Dis. 2022, 23, 253–261.
- Ishikawa, T.; Hayakawa, M.; Suzuki, H.; Ohno, E.; Mizutani, Y.; Iida, T.; Fujishiro, M.; Kawashima, H.; Hotta, K. Development of a Novel Evaluation Method for Endoscopic Ultrasound-Guided Fine-Needle Biopsy in Pancreatic Diseases Using Artificial Intelligence. Diagnostics 2022, 12, 434.
- Parasher, G.; Wong, M.; Rawat, M. Evolving role of artificial intelligence in gastrointestinal endoscopy. World J. Gastroenterol. 2020, 26, 7287–7298.
- Rodriguez, J.P.M.; Rodriguez, R.; Silva, V.W.K.; Kitamura, F.C.; Corradi, G.C.A.; de Marchi, A.C.B.; Rieder, R. Artificial intelligence as a tool for diagnosis in digital pathology whole slide images: A systematic review. J. Pathol. Inform. 2022, 13, 100138.
- Marya, N.B.; Powers, P.D.; Chari, S.T.; Gleeson, F.C.; Leggett, C.L.; Abu Dayyeh, B.K.; Chandrasekhara, V.; Iyer, P.G.; Majumder, S.; Pearson, R.K.; et al. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut 2021, 70, 1335–1344.
- Marya, N.B.; Powers, P.D.; Fujii-Lau, L.; Abu Dayyeh, B.K.; Gleeson, F.C.; Chen, S.; Long, Z.; Hough, D.M.; Chandrasekhara, V.; Iyer, P.G.; et al. Application of artificial intelligence using a novel EUS-based convolutional neural network model to identify and distinguish benign and malignant hepatic masses. Gastrointest. Endosc. 2021, 93, 1121–1130.e1.
- Oh, C.K.; Kim, T.; Cho, Y.K.; Cheung, D.Y.; Lee, B.-I.; Cho, Y.-S.; Kim, J.I.; Choi, M.-G.; Lee, H.H.; Lee, S. Convolutional neural network-based object detection model to identify gastrointestinal stromal tumors in endoscopic ultrasound images. J. Gastroenterol. Hepatol. 2021, 36, 3387–3394.
- Zhang, M.-M.; Yang, H.; Jin, Z.-D.; Yu, J.-G.; Cai, Z.-Y.; Li, Z.-S. Differential diagnosis of pancreatic cancer from normal tissue with digital imaging processing and pattern recognition based on a support vector machine of EUS images. Gastrointest. Endosc. 2010, 72, 978–985.
- Muhammad, W.; Hart, G.R.; Nartowt, B.; Farrell, J.J.; Johung, K.; Liang, Y.; Deng, J. Pancreatic Cancer Prediction Through an Artificial Neural Network. Front. Artif. Intell. 2019, 2, 2.
- Corral, J.E.; Hussein, S.; Kandel, P.; Bolan, C.W.; Bagci, U.; Wallace, M.B. Deep Learning to Classify Intraductal Papillary Mucinous Neoplasms Using Magnetic Resonance Imaging. Pancreas 2019, 48, 805–810.
- Cazacu, I.M.; Udristoiu, A.; Gruionu, L.G.; Iacob, A.; Gruionu, G.; Saftoiu, A. Artificial intelligence in pancreatic cancer: Toward precision diagnosis. Endosc. Ultrasound 2019, 8, 357–359.
- Săftoiu, A.; Vilmann, P.; Dietrich, C.F.; Iglesias-Garcia, J.; Hocke, M.; Seicean, A.; Ignee, A.; Hassan, H.; Streba, C.T.; Ioncică, A.M.; et al. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest. Endosc. 2015, 82, 59–69.
- Echle, A.; Rindtorff, N.T.; Brinker, T.J.; Luedde, T.; Pearson, A.T.; Kather, J.N. Deep learning in cancer pathology: A new generation of clinical biomarkers. Br. J. Cancer 2021, 124, 686–696.
- Kurita, Y.; Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Koda, H.; Tajika, M.; Shimizu, Y.; et al. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci. Rep. 2019, 9, 6893.
- Inoue, H.; Ogo, K.; Tabuchi, M.; Yamane, N.; Oka, H. An automatic visual inspection method based on supervised machine learning for rapid on-site evaluation in EUS-FNA. In Proceedings of the 2014 Proceedings of the SICE Annual Conference (SICE), Sapporo, Japan, 9–12 September 2014; pp. 1114–1119.
- Hashimoto, Y.; Ohno, I.; Imaoka, H.; Takahashi, H.; Mitsunaga, S.; Sasaki, M.; Kimura, G.; Suzuki, Y.; Watanabe, K.; Umemoto, K.; et al. Mo1296 Reliminary result of computer aided diagnosis (cad) performance using deep learning in eus-fna cytology of pancreatic cancer. Gastrointest. Endosc. 2018, 87, AB434.
This entry is offline, you can click here to edit this entry!
