Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Rare cells play essential roles in the initiation and progression of diseases and therefore their analysis is of great interest. The micro-magnetofluidic system is one of the emerging platforms that have been proposed for the rapid, sensitive, and cost-effective analysis of rare cells.
- microfluidics
- magnetic nanoparticles
- rare cell analysis
- cell sorting
1. Rare Cells: Definition and Examples
A cell is considered ‘rare’ when the number of cells of that subpopulation represents less than 0.01% of the total cell populations in a tissue or biofluid [1][2]. These cells, although occur as a minimal population, may play critical roles in disease progression and tissue regeneration. Below are a few examples highlighting the essentiality of rare cells in important bioprocesses.
For disease progression, one vivid instance is the initiation of cancer metastasis [3], where the primary tumor evolves and generates delayed secondary tumors in distal organs [4]. While many theories have been proposed to explain the process of cancer metastasis, the most accepted mechanism is through the formation of circulating tumor cells (CTCs) [5][6] (Figure 1A). In brief, CTCs are rare cells that emigrate from the primary tumor, enter and circulate within the peripheral blood, and eventually settle down in different organs for growing [7]. CTCs are generally considered as the seed of metastasis given their critical role during cancer migration [8][9] and therefore have attracted significant attention as a general biomarker for monitoring and predicting cancer progression [10][11][12]. However, one critical challenge of using CTCs is their rarity—the number of CTCs is extremely low, usually at the scale of a few CTCs in millions of normal cells [13][14][15].
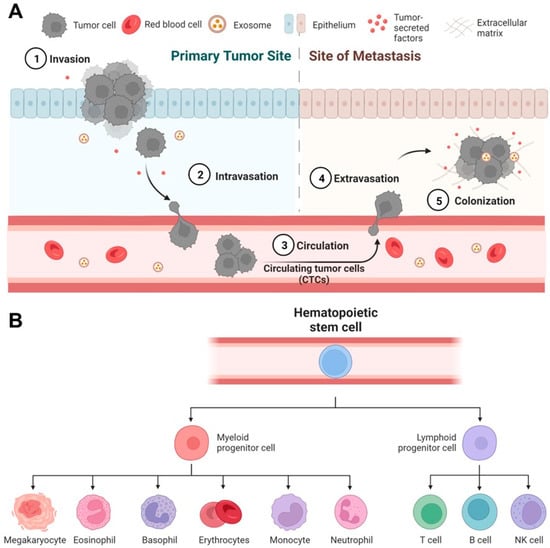
Figure 1. Rare cell-mediated bioprocesses in disease progression and tissue regeneration. (A) Circulating tumor cells (CTCs) play a critical role in cancer metastasis. In primary tumor sites, some highly invasive cancer cells undergo intravasation to enter the blood circulation and become CTCs. CTCs travel to distal organs and tissues, undergo extravasation, and eventually form deadly metastatic colonies. (B) Human hematopoietic stem/progenitor cells (HSPCs) have the potential to regenerate all types of blood cells. Under proper conditions in vitro and in vivo, HSPCs can differentiate into all myeloid (monocytes, neutrophils, etc.) and lymphoid cell lineages (T cells, B cells, etc.).
For tissue regeneration, a representative example is the use of human hematopoietic stem/progenitor cells (HSPCs) for transplantation. HSPCs only represent 0.2% to 0.5% of the leukocyte population (or 0.0003–0.0008% of the blood cell population) in peripheral blood [16], but have the full potency to produce all types of blood cells (e.g., lymphocytes and monocytes, Figure 1B). Therefore, the transplantation of HSPCs has been essential to the immune reconstruction post the treatment of leukemia [17][18][19]. Besides, rare cells may modulate the fate of a specific regenerative therapy given to a patient. It has been reported that rare undifferentiated stem cells in therapeutic cells, such as cardiomyocytes [20] and beta cells [21], may undergo uncontrolled growth in vivo in animal models [22][23] and eventually convert regenerative therapies into stem cell cancers [24][25][26][27].
Thus far, over 50 different types of microfluidic devices have been designed and validated for the analysis of rare cells, including circulating tumor cells [28][29], circulating tumor clusters [30][31][32], circulating cancer stem cells [33][34][35], circulating fetal cells [36][37], and circulating endothelial cells [38] in peripheral blood, as well as contaminating tumor cells [39] in therapeutic cell products. Multiple strategies have been proposed, such as filtration, inertial fluidics, deterministic lateral displacement (DLD), and immunomagnetic sorting [40].
2. Micro-Magnetic Deflection
2.1. Principles
Magnetic deflection is a main approach of magnetophoresis. It relies on the dictated movement/migration of target cells to achieve separation. The main procedures of magnetic deflection include the specific magnetic labeling of target cells, the incorporation of an external magnetic field, and on-chip magnetic sorting.
The magnetic deflection of target cells is the result of interaction between magnetic force and hydrodynamic force on the cells. In the presence of an external magnetic field, magnetically labeled cells experience a magnetic force produced by the inserted magnetic gradient. In the meantime, the cells experience Stoke’s drag force from the fluid flow. The magnetic force given as
where 𝑛 is the number of magnetic beads bound to the target cell, 𝑉𝑏𝑒𝑎𝑑 is the volume of a nanobead, ∆𝜒𝑏𝑒𝑎𝑑∆ is the relative magnetic susceptibility of nanobeads, 𝜇0 is the magnetic permeability of the free space, and 𝑩 is the magnetic field.
The drag force is expressed as
where 𝜂 is the viscosity of the ambient fluid, 𝑅 is the radius of the t cell, and 𝑈 is the velocity magnitude of the target cells relative to the flow.
To dictate magnetic deflection, both magnetic force and hydrodynamic force can be designed based on applications. For magnetic force manipulation, the magnetic labeling of target cells and magnetic gradient in the microchannel can be designed. The optimization of magnetic labeling was discussed above. The change in magnetic gradients can be achieved by modifying the magnetic field. For the optimization of hydrodynamic force, the flow pattern in the microchannel can be designed.
2.2. Magnetic and Hydrodynamic Optimization
The design of magnetic field is an important aspect for the optimization of magnetic deflection. To achieve a good deflection, a large magnetic gradient is required. To increase the magnetic gradient, researchers have tried different types of permanent magnets and different ways to place the magnets. The easiest strategy is to apply a macroscale permanent magnet on the side or at the bottom of the microfluidic device to produce magnetic gradient [41][42][43]. Target cells will be deflected due to the non-uniform magnetic field produced by the permanent magnet. However, the magnetic gradient may not be strong enough to achieve high-resolution magnetic deflection when target cells have low magnetic susceptibility. Alternatively, magnetic blocks arranged with opposing poles were used for a larger magnetic field gradient [44][45]. Micro-magnet arrays have also been applied to create strong local magnetic gradients to increase magnetic deflection sensitivity [46][47].
Insertion of paramagnetic materials has also been used to further enhance local magnetic gradients in the microfluidic device. Soft magnetic materials such as nickel and ion oxide micro- or nanostructures were integrated in microfluidic devices to create strong local magnetic gradients in the microchannel [48][49][50][51]. Tom Soh et al. have designed nickel microchannel strips to deflect magnetic labeled rare cells [52]. The magnetic ratcheting cytometry system developed by the Di Carlo group also applied soft magnetic materials for magnetic field enhancement and adjustment. In this system, micropillars made of soft materials were arranged at various distances to create multiple magnetic ratcheting zones in the sorting chamber. Periodically switching magnetic fields were produced under the microchip through rotating permanent magnets. The micropillars activated by the switching magnetic field drove magnetically labeled cells across the sorting chamber, and cells with different magnetic loading levels would be stabilized at different ratcheting zones [53][54]. In recent years, the Kelley group has developed a magnetic deflection-based microfluidic chip-termed PRISM [55][56][57]. This system made use of the superb ferromagnetic property of an amorphous metal alloy ribbon called metglas to produce an extremely strong local magnetic field when external magnets were introduced. Through lithography and wet etching, metglas-based magnetic guide could be patterned on a glass substrate. SU8-based microchannels were then generated on top of the magnetic guide for sample processing and magnetic deflection. The magnetic guides embedded under the microchannel were designed to branch out from the middle to the sidewall of the microchannel with multiple magnetic deflection angles. Magnetically labeled rare cells were thus segregated into different subpopulations when following the magnetic guides.
Making use of hydrodynamic force is another way to dictate magnetic deflection. For example, designing microstructures in the microchannel to produce a low drag force zone can be used to isolate magnetically labeled cells for single-cell analysis [58]. Using different materials for channel substrates and optimized channel shapes to obtain uniform flow profiles is another way for drag force manipulation [59]. A third way is to introduce ‘flow pockets’ on the side the microchannel and apply permanent magnets along the ‘flow pockets’ [60][61]. The flow velocity is close to zero in the ‘flow pockets’ and non-target cells flowing along the fluid flow will not enter the ‘flow pockets’, while magnetic labeled target cells will be deflected and trapped in the ‘flow pockets’. Table 1 summarizes different magnetic sorting methods and their relevant performance including throughput, target cell isolation efficiency, purity, sensitivity and their potential limitations.
Due to the miniaturized channel scale and strong inserted external magnetic field, microfluidics-based magnetic deflection can bring high deflection efficiency of target cells with good purity [62][63][64][65]. Additionally, since magnetically labeled cells move continuously along the sorting chamber, deflected target cells can be easily collected in the designated outlet with few cell loss. However, this method solely depends on magnetic force added on the target cells, which may not be enough to address the heterogeneity of target cells. Rare cells such as CTCs are known to be heterogenous in both biological and physical properties [66][67]. For example, CTCs undergoing epithelial to mesenchymal transition (EMT) will lose the expression of EpCAM [68][69][70][71]. Targeting EpCAM alone cannot isolate CTCs with low EpCAM expression. To address this problem, researchers introduced multiplex marker targeting [72][73][74]. In addition to EpCAM, biomarkers such N-cadherin, vimentin were used to differentiate CTCs under EMT from blood cells [75][76]. Alternatively, magnetic deflection can be combined with physical-property based cell separation methods to achieve superb CTC isolation efficiency.
Table 1. Comparison of different magnetic deflection-based cell sorting strategies.
| Sorting Strategy | Application | Throughput | Efficiency | Purity | Sensitivity | Limitation | Reference |
|---|---|---|---|---|---|---|---|
| Magnetic guided deflection | CTC and CTC cluster isolation | 0.5 mL/h | ~90% | 5.7 log white blood cell depletion | 10 targe cell per mL | Strong cell–cell interaction at high cell concentration can affect its performance | [56] |
| Modified magnetic beads | Separation of cancer cells | NA | ~90% | >80% | 106 cells /mL | Large bead size reduce sensitivity | [77] |
| Captured magnetic bead-bounded target cells with dead-ended side chambers near the a permanent magnet | CTC isolation from whole blood | 1.2 mL/h | >90% | <0.4% white blood cell capture | 2–80 target cells spiked in 1 mL of blood | Lack of clinical sample processing | [42] |
| Magnetophoresis assisted cell capture activated by magnet arrays | Isolation of cancer cells spiked in human blood | 7.2 mL/h | >60% | ~30% | 3.5 × 104 cancer cells spiked in 1 mL of blood | Release of the rare cells would be challenging | [44] |
| Two-stage magnetic separation | White blood cell sorting from whole blood | 1.2 mL/h | 93% | >70% | 103 cells/min | Require further optimization of rare cell separation | [49] |
| Ferromagnetic guide based deflection in a large scale 3D printed system | Isolation of mature natural killer cells from blood | >18 mL/h | >50% | ~90% | 107 cells per mL | Complicated fabrication and assembly process | [57] |
| Magnetic deflection facilitated single-cell capture | Capture of rare tumor cells from mouse blood | 2 mL/h | ~90% | >90% | <100 CTCs from 1 mL of whole blood | The system can be saturated due to the limited number of trap units | [58] |
| Magnetic deflection and capture on wavy-herringbone structures | Capture CTCs from whole blood | 0.54 mL/h | 92% | 91% | 100 target cells per mL blood | Lack of clinical processing | [61] |
| Magnetic ratcheting | Profiling of magnetically labeled immune cells in spiked samples | NA | 87% | 95% | 107 cells per mL | The throughput was limited as no in-flow allowed during magnetic ratcheting | [53] |
2.3. Fabrication of Magnetic Deflection-Based Microfluidic Devices
The functioning of magnetic deflection-based microfluidic devices requires two main components: microchannel/sorting chamber assembly and magnetic component combination. For most designs, the sorting chamber for magnetic deflection is integrated to the microchannels for sampling. Polydimethylsiloxane (PDMS) is the most common material for channel fabrication as it is transparent, air permeable, and biocompatible. PDMS substrates with microchannel/sorting chamber features can be conveniently fabricated using soft lithography [78]. Briefly, a mold such as a silicon master containing the channel features can be first fabricated using photolithography, and then liquid PDMS mix (base: curing agent = 10:1) is casted on the mold and polymerized at 70 °C for 2 h. The PDMS substrate is then bonded with a cover (e.g., a thin glass slide) to form the microchannel. Other than PDMS, other materials such as negative photoresist (e.g., SU8) were also used in channel fabrication [56]. SU8 channels can be fabricated through photolithography. For the integration of magnetic components, it can be direct integration by simply applying a permanent magnet at the top/bottom or on the side of the microchannel. It can also be a complicated process by adding soft magnetic materials. For the insertion of soft magnetic materials, there are multiple ways. The soft magnetic material can first be mixed in solution and then introduced to the bottom substrate of the device through injection molding [48]. Another way is to use wet etching to generate magnetic deflection features. For this method, the soft magnetic materials are first coated on the device substrate through electron beam physical vapor deposition or epoxy taping [56][79]. The soft magnetic layer is then patterned through wet etching masked by a photoresist layer. A third way to generate soft magnetic features is through electroplating [53]. Different fabrications have pros and cons. For example, liquid soft magnetic material injection can plate magnetic materials easily, but it requires a mold with mirochannels for injection. Therefore, the shape and size of soft magnetic features can be limited. Deposited magnetic features can be highly accurate, but the thickness and magnetic strength of the material can be limited. For the taping of ferromagnetic material such as metglas, it can introduce an extremely strong local magnetic field. However, the taping process limits the smoothness of the substrate surface. Electroplated magnetic features can be generated with relatively high aspect ratio and brings strong local magnetic field. However, the resolution of electroplated feature is relatively low. As such, the fabrication methods chosen usually depends on the specific application of the microfluidic device.
2.4. Integration and Applications for Rare Cell Capture
Different approaches have been reported to combine magnetic deflection with other cell separation mechanisms, showing high compatibility of magnetic deflection. The integration of label-free cell sorting methods and magnetic deflection was widely explored since it usually shows advantages of both approaches. Label-free cell sorting methods generally give high throughput, while magnetic deflection shows high specificity. Toner et al. have developed an integrated microfluidic system that includes deterministic lateral displacement (DLD), inertial focusing and magnetic separation [80]. This system was able to isolate CTCs from untreated whole blood, where nucleated cells were first separated from red blood cells (RBCs) through DLD, followed by magnetic depletion of magnetically labeled white blood cells (WBCs). Since RBCs are significantly smaller than nucleated cells, DLD can separate nucleated cells from RBCs with high purity. Inertial focusing re-aligns nucleated cell mixture into single-cell strips which reduces the cell–cell interaction. Therefore, in the last step, the magnetic depletion of WBCs brought high CTC efficiency and purity. Similarly, Nasiri et al. presented a hybrid device combing inertial focusing and magnetic deflection for CTC isolation [81]. High isolation efficiency and purity can also be achieved. Several groups have combined dielectrophoresis (DEP) with magnetic separation for rare cell separation [82][83][84]. Nucleated cells were separated by size using DEP and target cells were then isolated through magnetic deflection. Combing magnetic deflection with acoustic-based sorting for the isolation of rare cells from whole blood was also widely studied [85]. Acoustics is a technology used for separating particles/cells based on the acoustic force which is proportional to the size of cells. As such, acoustics can also be used for the pre-sort of nucleated cells when integrated with magnetic deflection. Several other mechanisms incorporated with magnetic deflection were also reported to be highly efficient. For instance, Alipanah et al. developed a rare cell sorter using induced charged electroosmotic flow and magnetophoresis [86]. Shamloo et al. presented a system combining centrifugal force with magnetic deflection for efficiency and high-purity isolation of rare cells [87]. Javanmard et al. described a novel approach for the rapid assessment of surface markers on cancer cells using a combination of immunomagnetic separation and multi-frequency impedance cytometry. In this work, multi-frequency impedance cytometry was applied for target cell detection rather than separation [88].
This entry is adapted from the peer-reviewed paper 10.3390/chemosensors11060335
References
- Proserpio, V.; Lonnberg, T. Single-cell technologies are revolutionizing the approach to rare cells. Immunol. Cell Biol. 2016, 94, 225–229.
- Dharmasiri, U.; Witek, M.A.; Adams, A.A.; Soper, S.A. Microsystems for the capture of low-abundance cells. Annu. Rev. Anal. Chem. 2010, 3, 409–431.
- Maheswaran, S.; Haber, D.A. Circulating tumor cells: A window into cancer biology and metastasis. Curr. Opin. Genet. Dev. 2010, 20, 96–99.
- Zhang, J.; Chen, K.; Fan, Z.H. Circulating Tumor Cell Isolation and Analysis. Adv. Clin. Chem. 2016, 75, 1–31.
- Poudineh, M.; Sargent, E.H.; Pantel, K.; Kelley, S.O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng. 2018, 2, 72–84.
- Abouali, H.; Hosseini, S.A.; Purcell, E.; Nagrath, S.; Poudineh, M. Recent Advances in Device Engineering and Computational Analysis for Characterization of Cell-Released Cancer Biomarkers. Cancers 2022, 14, 288.
- Green, B.J.; Saberi Safaei, T.; Mepham, A.; Labib, M.; Mohamadi, R.M.; Kelley, S.O. Beyond the Capture of Circulating Tumor Cells: Next-Generation Devices and Materials. Angew. Chem. Int. Ed. Engl. 2016, 55, 1252–1265.
- Pantel, K.; Speicher, M.R. The biology of circulating tumor cells. Oncogene 2016, 35, 1216–1224.
- Woo, D.; Yu, M. Circulating tumor cells as “liquid biopsies” to understand cancer metastasis. Transl. Res. 2018, 201, 128–135.
- Alva, A.; Friedlander, T.; Clark, M.; Huebner, T.; Daignault, S.; Hussain, M.; Lee, C.; Hafez, K.; Hollenbeck, B.; Weizer, A.; et al. Circulating Tumor Cells as Potential Biomarkers in Bladder Cancer. J. Urol. 2015, 194, 790–798.
- Danila, D.C.; Fleisher, M.; Scher, H.I. Circulating tumor cells as biomarkers in prostate cancer. Clin. Cancer Res. 2011, 17, 3903–3912.
- Yap, T.A.; Lorente, D.; Omlin, A.; Olmos, D.; de Bono, J.S. Circulating tumor cells: A multifunctional biomarker. Clin. Cancer Res. 2014, 20, 2553–2568.
- den Toonder, J. Circulating tumor cells: The Grand Challenge. Lab Chip 2011, 11, 375–377.
- Yang, Y.P.; Giret, T.M.; Cote, R.J. Circulating Tumor Cells from Enumeration to Analysis: Current Challenges and Future Opportunities. Cancers 2021, 13, 2723.
- Andree, K.C.; van Dalum, G.; Terstappen, L.W. Challenges in circulating tumor cell detection by the CellSearch system. Mol. Oncol. 2016, 10, 395–407.
- Lemos, N.E.; Farias, M.G.; Kubaski, F.; Scotti, L.; Onsten, T.G.H.; Brondani, L.A.; Wagner, S.C.; Sekine, L. Quantification of peripheral blood CD34(+) cells prior to stem cell harvesting by leukapheresis: A single center experience. Hematol. Transfus. Cell Ther. 2018, 40, 213–218.
- Park, B.; Yoo, K.H.; Kim, C. Hematopoietic stem cell expansion and generation: The ways to make a breakthrough. Blood Res. 2015, 50, 194–203.
- Marquez-Curtis, L.A.; Turner, A.R.; Sridharan, S.; Ratajczak, M.Z.; Janowska-Wieczorek, A. The ins and outs of hematopoietic stem cells: Studies to improve transplantation outcomes. Stem Cell Rev. Rep. 2011, 7, 590–607.
- Rajendiran, S.; Boyer, S.W.; Forsberg, E.C. A quantitative hematopoietic stem cell reconstitution protocol: Accounting for recipient variability, tissue distribution and cell half-lives. Stem Cell Res. 2020, 50, 102145.
- Ben-David, U.; Gan, Q.F.; Golan-Lev, T.; Arora, P.; Yanuka, O.; Oren, Y.S.; Leikin-Frenkel, A.; Graf, M.; Garippa, R.; Boehringer, M.; et al. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell 2013, 12, 167–179.
- Han, L.; He, H.; Yang, Y.; Meng, Q.; Ye, F.; Chen, G.; Zhang, J. Distinctive Clinical and Pathologic Features of Immature Teratomas Arising from Induced Pluripotent Stem Cell-Derived Beta Cell Injection in a Diabetes Patient. Stem Cells Dev. 2022, 31, 97–101.
- Hong, S.G.; Winkler, T.; Wu, C.; Guo, V.; Pittaluga, S.; Nicolae, A.; Donahue, R.E.; Metzger, M.E.; Price, S.D.; Uchida, N.; et al. Path to the clinic: Assessment of iPSC-based cell therapies in vivo in a nonhuman primate model. Cell Rep. 2014, 7, 1298–1309.
- Muller, F.J.; Goldmann, J.; Loser, P.; Loring, J.F. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell 2010, 6, 412–414.
- Fong, C.Y.; Gauthaman, K.; Bongso, A. Teratomas from pluripotent stem cells: A clinical hurdle. J. Cell Biochem. 2010, 111, 769–781.
- Blum, B.; Benvenisty, N. The tumorigenicity of human embryonic stem cells. Adv. Cancer Res. 2008, 100, 133–158.
- Lee, A.S.; Tang, C.; Rao, M.S.; Weissman, I.L.; Wu, J.C. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 2013, 19, 998–1004.
- Wellmerling, K.; Lehmann, C.; Singh, A.; Kirby, B.J. Microfluidic chip for label-free removal of teratoma-forming cells from therapeutic human stem cells. J. Immunol. Regen. Med. 2020, 10, 100030.
- Chen, K.; Georgiev, T.Z.; Sheng, W.; Zheng, X.; Varillas, J.I.; Zhang, J.; Hugh Fan, Z. Tumor cell capture patterns around aptamer-immobilized microposts in microfluidic devices. Biomicrofluidics 2017, 11, 054110.
- Chen, K.; Dopico, P.; Varillas, J.; Zhang, J.; George, T.J.; Fan, Z.H. Integration of Lateral Filter Arrays with Immunoaffinity for Circulating-Tumor-Cell Isolation. Angew. Chem. Int. Ed. Engl. 2019, 58, 7606–7610.
- Green, B.J.; Marazzini, M.; Hershey, B.; Fardin, A.; Li, Q.; Wang, Z.; Giangreco, G.; Pisati, F.; Marchesi, S.; Disanza, A.; et al. PillarX: A Microfluidic Device to Profile Circulating Tumor Cell Clusters Based on Geometry, Deformability, and Epithelial State. Small 2022, 18, e2106097.
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691.
- Au, S.H.; Edd, J.; Stoddard, A.E.; Wong, K.H.K.; Fachin, F.; Maheswaran, S.; Haber, D.A.; Stott, S.L.; Kapur, R.; Toner, M. Microfluidic Isolation of Circulating Tumor Cell Clusters by Size and Asymmetry. Sci. Rep. 2017, 7, 2433.
- Varillas, J.I.; Zhang, J.; Chen, K.; Barnes, I.I.; Liu, C.; George, T.J.; Fan, Z.H. Microfluidic Isolation of Circulating Tumor Cells and Cancer Stem-Like Cells from Patients with Pancreatic Ductal Adenocarcinoma. Theranostics 2019, 9, 1417–1425.
- Cho, H.Y.; Choi, J.H.; Lim, J.; Lee, S.N.; Choi, J.W. Microfluidic Chip-Based Cancer Diagnosis and Prediction of Relapse by Detecting Circulating Tumor Cells and Circulating Cancer Stem Cells. Cancers 2021, 13, 1385.
- Obermayr, E.; Koppensteiner, N.; Heinzl, N.; Schuster, E.; Holzer, B.; Fabikan, H.; Weinlinger, C.; Illini, O.; Hochmair, M.; Zeillinger, R. Cancer Stem Cell-Like Circulating Tumor Cells Are Prognostic in Non-Small Cell Lung Cancer. J. Pers. Med. 2021, 11, 1225.
- Zhang, H.; Yang, Y.; Liu, Y.; Wang, Y.; Ruan, W.; Song, J.; Yu, X.; Wu, L.; Zhu, Z.; Hong, G.; et al. Stimuli-Responsive Microfluidic Interface Enables Highly Efficient Capture and Release of Circulating Fetal Cells for Non-Invasive Prenatal Testing. Anal. Chem. 2020, 92, 9281–9286.
- Winter, M.; Hardy, T.; Rezaei, M.; Nguyen, V.; Zander-Fox, D.; Ebrahimi Warkiani, M.; Thierry, B. Isolation of Circulating Fetal Trophoblasts Using Inertial Microfluidics for Noninvasive Prenatal Testing. Adv. Mater. Technol. 2018, 3, 1800066.
- Chen, S.; Sun, Y.; Neoh, K.H.; Chen, A.; Li, W.; Yang, X.; Han, R.P.S. Microfluidic assay of circulating endothelial cells in coronary artery disease patients with angina pectoris. PLoS ONE 2017, 12, e0181249.
- Wang, Z.; Sargent, E.H.; Kelley, S.O. Ultrasensitive Detection and Depletion of Rare Leukemic B Cells in T Cell Populations via Immunomagnetic Cell Ranking. Anal. Chem. 2021, 93, 2327–2335.
- Chen, Y.; Li, P.; Huang, P.H.; Xie, Y.; Mai, J.D.; Wang, L.; Nguyen, N.T.; Huang, T.J. Rare cell isolation and analysis in microfluidics. Lab Chip 2014, 14, 626–645.
- Zhao, W.; Cheng, R.; Lim, S.H.; Miller, J.R.; Zhang, W.; Tang, W.; Xie, J.; Mao, L. Biocompatible and label-free separation of cancer cells from cell culture lines from white blood cells in ferrofluids. Lab Chip 2017, 17, 2243–2255.
- Kang, J.H.; Krause, S.; Tobin, H.; Mammoto, A.; Kanapathipillai, M.; Ingber, D.E. A combined micromagnetic-microfluidic device for rapid capture and culture of rare circulating tumor cells. Lab Chip 2012, 12, 2175–2181.
- Zhao, W.; Zhu, T.; Cheng, R.; Liu, Y.; He, J.; Qiu, H.; Wang, L.; Nagy, T.; Querec, T.D.; Unger, E.R.; et al. Label-Free and Continuous-Flow Ferrohydrodynamic Separation of HeLa Cells and Blood Cells in Biocompatible Ferrofluids. Adv. Funct. Mater. 2016, 26, 3990–3998.
- Unni, M.; Zhang, J.L.; George, T.J.; Segal, M.S.; Fan, Z.H.; Rinaldi, C. Engineering magnetic nanoparticles and their integration with microfluidics for cell isolation. J. Colloid Interface Sci. 2020, 564, 204–215.
- Hoshino, K.; Huang, Y.-Y.; Lane, N.; Huebschman, M.; Uhr, J.W.; Frenkel, E.P.; Zhang, X. Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 2011, 11, 3449–3457.
- Labib, M.; Wang, Z.; Ahmed, S.U.; Mohamadi, R.M.; Duong, B.; Green, B.; Sargent, E.H.; Kelley, S.O. Tracking the expression of therapeutic protein targets in rare cells by antibody-mediated nanoparticle labelling and magnetic sorting. Nat. Biomed. Eng. 2021, 5, 41–52.
- Khojah, R.; Xiao, Z.; Panduranga, M.K.; Bogumil, M.; Wang, Y.; Goiriena-Goikoetxea, M.; Chopdekar, R.V.; Bokor, J.; Carman, G.P.; Candler, R.N.; et al. Single-Domain Multiferroic Array-Addressable Terfenol-D (SMArT) Micromagnets for Programmable Single-Cell Capture and Release. Adv. Mater. 2021, 33, e2006651.
- Royet, D.; Heriveaux, Y.; Marchalot, J.; Scorretti, R.; Dias, A.; Dempsey, N.M.; Bonfim, M.; Simonet, P.; Frenea-Robin, M. Using injection molding and reversible bonding for easy fabrication of magnetic cell trapping and sorting devices. J. Magn. Magn. Mater. 2017, 427, 306–313.
- Lin, S.; Zhi, X.; Chen, D.; Xia, F.; Shen, Y.; Niu, J.; Huang, S.; Song, J.; Miao, J.; Cui, D.; et al. A flyover style microfluidic chip for highly purified magnetic cell separation. Biosens. Bioelectron. 2019, 129, 175–181.
- Yu, X.; Feng, X.; Hu, J.; Zhang, Z.L.; Pang, D.W. Controlling the magnetic field distribution on the micrometer scale and generation of magnetic bead patterns for microfluidic applications. Langmuir ACS J. Surf. Colloids 2011, 27, 5147–5156.
- Inglis, D.W.; Riehn, R.; Austin, R.H.; Sturm, J.C. Continuous microfluidic immunomagnetic cell separation. Appl. Phys. Lett. 2004, 85, 5093–5095.
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994.
- Murray, C.; Miwa, H.; Dhar, M.; Park, D.E.; Pao, E.; Martinez, J.; Kaanumale, S.; Loghin, E.; Graf, J.; Rhaddassi, K. Unsupervised capture and profiling of rare immune cells using multi-directional magnetic ratcheting. Lab Chip 2018, 18, 2396–2409.
- Adeyiga, O.B.; Murray, C.; Muñoz, H.E.; Escobar, A.; Di Carlo, D. Magnetic microparticle concentration and collection using a mechatronic magnetic ratcheting system. PLoS ONE 2021, 16, e0246124.
- Mair, B.; Aldridge, P.M.; Atwal, R.S.; Philpott, D.; Zhang, M.; Masud, S.N.; Labib, M.; Tong, A.H.Y.; Sargent, E.H.; Angers, S.; et al. High-throughput genome-wide phenotypic screening via immunomagnetic cell sorting. Nat. Biomed. Eng. 2019, 3, 796–805.
- Aldridge, P.M.; Mukhopadhyay, M.; Ahmed, S.U.; Zhou, W.; Christinck, E.; Makonnen, R.; Sargent, E.H.; Kelley, S.O. Prismatic Deflection of Live Tumor Cells and Cell Clusters. ACS Nano 2018, 12, 12692–12700.
- Philpott, D.N.; Chen, K.; Atwal, R.S.; Li, D.; Christie, J.; Sargent, E.H.; Kelley, S.O. Ultrathroughput immunomagnetic cell sorting platform. Lab Chip 2022, 22, 4822–4830.
- Ma, Y.; Chen, K.; Xia, F.; Atwal, R.; Wang, H.; Ahmed, S.U.; Cardarelli, L.; Lui, I.; Duong, B.; Wang, Z.; et al. Phage-Based Profiling of Rare Single Cells Using Nanoparticle-Directed Capture. ACS Nano 2021, 15, 19202–19210.
- Fernandez, C.G.; Pastora, J.G.; Basauri, A.; Fallanza, M.; Bringas, E.; Chalmers, J.J.; Ortiz, I. Continuous-Flow Separation of Magnetic Particles from Biofluids: How Does the Microdevice Geometry Determine the Separation Performance? Sensors 2020, 20, 3030.
- Huang, N.T.; Hwong, Y.J.; Lai, R.L. A microfluidic microwell device for immunomagnetic single-cell trapping. Microfluid. Nanofluid. 2018, 22, 16.
- Shi, W.; Wang, S.; Maarouf, A.; Uhl, C.G.; He, R.; Yunus, D.; Liu, Y. Magnetic particles assisted capture and release of rare circulating tumor cells using wavy-herringbone structured microfluidic devices. Lab Chip 2017, 17, 3291–3299.
- Wyatt Shields, C., IV; Reyes, C.D.; López, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249.
- Shen, Y.; Yalikun, Y.; Tanaka, Y. Recent advances in microfluidic cell sorting systems. Sens. Actuators B Chem. 2019, 282, 268–281.
- Yu, Z.T.F.; Aw Yong, K.M.; Fu, J. Microfluidic Blood Cell Sorting: Now and Beyond. Small 2014, 10, 1687–1703.
- Yun, H.; Kim, K.; Lee, W.G. Cell manipulation in microfluidics. Biofabrication 2013, 5, 022001.
- Marrinucci, D.; Bethel, K.; Kolatkar, A.; Luttgen, M.S.; Malchiodi, M.; Baehring, F.; Voigt, K.; Lazar, D.; Nieva, J.; Bazhenova, L.; et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys. Biol. 2012, 9, 016003.
- Lang, J.M.; Casavant, B.P.; Beebe, D.J. Circulating Tumor Cells: Getting More from Less. Sci. Transl. Med. 2012, 4, 141ps13.
- Hyun, K.A.; Koo, G.B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.I.; Jung, H.I.; Kim, Y.S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016, 7, 24677–24687.
- Sankpal, N.V.; Fleming, T.P.; Sharma, P.K.; Wiedner, H.J.; Gillanders, W.E. A double-negative feedback loop between EpCAM and ERK contributes to the regulation of epithelial–mesenchymal transition in cancer. Oncogene 2017, 36, 3706–3717.
- Keller, L.; Werner, S.; Pantel, K. Biology and clinical relevance of EpCAM. Cell Stress 2019, 3, 165–180.
- Tellez-Gabriel, M.; Heymann, M.-F.; Heymann, D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics 2019, 9, 4580–4594.
- Issadore, D.; Chung, J.; Shao, H.; Liong, M.; Ghazani, A.A.; Castro, C.M.; Weissleder, R.; Lee, H. Ultrasensitive Clinical Enumeration of Rare Cells ex vivo Using a Micro-Hall Detector. Sci. Transl. Med. 2012, 4, 141ra92.
- Scharpenseel, H.; Hanssen, A.; Loges, S.; Mohme, M.; Bernreuther, C.; Peine, S.; Lamszus, K.; Goy, Y.; Petersen, C.; Westphal, M.; et al. EGFR and HER3 expression in circulating tumor cells and tumor tissue from non-small cell lung cancer patients. Sci. Rep. 2019, 9, 7406.
- Broersen, L.H.A.; van Pelt, G.W.; Tollenaar, R.A.E.M.; Mesker, W.E. Clinical application of circulating tumor cells in breast cancer. Cell. Oncol. 2014, 37, 9–15.
- Menyailo, M.E.; Tretyakova, M.S.; Denisov, E.V. Heterogeneity of Circulating Tumor Cells in Breast Cancer: Identifying Metastatic Seeds. Int. J. Mol. Sci. 2020, 21, 1696.
- Jie, X.-X.; Zhang, X.-Y.; Xu, C.-J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: Mechanisms and clinical applications. Oncotarget 2017, 8, 81558.
- Yin, D.; Shi, A.; Zhou, B.; Wang, M.; Xu, G.; Shen, M.; Zhu, X.; Shi, X. Efficient Capture and Separation of Cancer Cells Using Hyaluronic Acid-Modified Magnetic Beads in a Microfluidic Chip. Langmuir ACS J. Surf. Colloids 2022, 38, 11080–11086.
- Qin, D.; Xia, Y.; Whitesides, G.M. Soft lithography for micro- and nanoscale patterning. Nat. Protoc. 2010, 5, 491–502.
- Adams, J.D.; Kim, U.; Soh, H.T. Multitarget magnetic activated cell sorter. Proc. Natl. Acad. Sci. USA 2008, 105, 18165–18170.
- Karabacak, N.M.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.I.; et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2014, 9, 694–710.
- Nasiri, R.; Shamloo, A.; Akbari, J. Design of a Hybrid Inertial and Magnetophoretic Microfluidic Device for CTCs Separation from Blood. Micromachines 2021, 12, 877.
- Cetin, B.; Ozer, M.B.; Cagatay, E.; Buyukkocak, S. An integrated acoustic and dielectrophoretic particle manipulation in a microfluidic device for particle wash and separation fabricated by mechanical machining. Biomicrofluidics 2016, 10, 014112.
- Krishnan, J.N.; Kim, C.; Park, H.J.; Kang, J.Y.; Kim, T.S.; Kim, S.K. Rapid microfluidic separation of magnetic beads through dielectrophoresis and magnetophoresis. Electrophoresis 2009, 30, 1457–1463.
- Kim, U.; Soh, H.T. Simultaneous sorting of multiple bacterial targets using integrated Dielectrophoretic-Magnetic Activated Cell Sorter. Lab Chip 2009, 9, 2313–2318.
- Adams, J.D.; Thevoz, P.; Bruus, H.; Soh, H.T. Integrated acoustic and magnetic separation in microfluidic channels. Appl. Phys. Lett. 2009, 95, 254103.
- Alipanah, M.; Hafttananian, M.; Hedayati, N.; Ramiar, A.; Alipanah, M. Microfluidic on-demand particle separation using induced charged electroosmotic flow and magnetic field. J. Magn. Magn. Mater. 2021, 537, 168156.
- Shamloo, A.; Naghdloo, A.; Besanjideh, M. Cancer cell enrichment on a centrifugal microfluidic platform using hydrodynamic and magnetophoretic techniques. Sci. Rep. 2021, 11, 1939.
- Lin, Z.; Lin, S.-Y.; Xie, P.; Lin, C.-Y.; Rather, G.M.; Bertino, J.R.; Javanmard, M. Rapid assessment of surface markers on cancer cells using immuno-magnetic separation and multi-frequency impedance cytometry for targeted therapy. Sci. Rep. 2020, 10, 3015.
This entry is offline, you can click here to edit this entry!
