Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Horticulture
Salinity stress is a major threat to the commonly grown crop cultivars. The close relatives of modern cultivated plants such as their crop wild relatives (CWRs), can be a promising and sustainable resource to broaden the diversity of crops for the salinity stress tolerances. Contemporary developments in the transcriptomic technologies have revealed the valuable genetic variation of CWRs that represents a practical gene pool for improving the plant’s adaptability to salt stress.
- salinity
- TFs
- CWRs
- transcriptomics
- stress tolerance
1. Introduction
The climatic changes and current plant cultivating practices of poor land management and have pressurized the agri-food systems with salinity stress. These activities have resulted in the salinization of 20% of the irrigated lands, which incurs a major part of the annual agricultural losses [1]. Such salt stress issues are detrimental to crop production of the South Asian, Middle Eastern, and sub-Saharan African regions where agricultural production is limited the most, and can lead to persistence of food scarcity around the world [2]. This shows that salinity stress is a major obstacle to present and future global food production. Hence, addressing the issue of salinity will provide a compelling contribution to the economy of producers and to attaining the food demands needed to sustain the 10 billion people by 2050. This clearly makes it important to identify ways to alleviate the effects of salt stress by using saline soils and water to expand agricultural yield.
It is essential to breed or engineer novel crop varieties that are tolerant to salinity for utilizing the salt-affected lands, which can contribute to the sustainable intensification of agronomic systems. The degree of salt tolerance differs among plant species based on their genetic variations. Accordingly, all the available genetic diversity needs to be exploited and the current gene pools of the cultivated plants need to be expanded. One of the possible ways to attain this solution is to explore the sources of wild desirable genes from the crop wild relatives (CWRs) [3]. These crops are the ancestors or progenitors of domesticated plants that have higher genetic and phenotypic variability. CWRs are present in their natural habitats or near their centers of origin. Unlike the domesticated species, the wild relatives were not subjected to human selection pressure. Instead, they developed and adapted in abundance throughout their evolutionary history and kept evolving in harsh climatic conditions including salinity. CWRs consist of a plethora of genes that are involved in their enhanced tolerance to salt stress, which represents a rich pool of alleles that are absent in the current cultivars with crucial agronomic value [4]. These resources when utilized are likely to broaden the genetic base of cultivated crops by introducing economically important genes, critical for meeting the challenges of food crisis and salinity stress.
Salt stress is a complicated phenomenon during which the CWRs have developed numerous adaptive responses, morphologically, physiologically, and biochemically. These responses are controlled at the molecular level by a large number of genes that signal, synthesize phytohormones, transport ions, and cause morphological changes [5]. To decipher these molecular mechanisms, transcriptomics is a powerful method to be employed. This will ascertain their use for future food security, which has not been clearly recognized despite their capability as gene donors for crop enhancement. The combination of CWR breeding with the latest biotechnological advancements such as transcriptomics can be widely utilized for successfully generating salt-stress-tolerant plants. This is because coupling CWRs transcriptomics with in silico analysis can lead to an unbiased transcriptomic profile development for the identification and activation of certain physiological mechanisms, set of genes, and specified genetic pathways involved in plant salt stress tolerance.
2. Impact of Salinity Stress on Plants and its Adaptative Mechanism
Salt stress is a complex abiotic stress that substantially affects the plant’s productivity. It occurs largely due to the high concentration of soluble salt ions within the soil solution that generates different stresses within the plants. It lowers the soil water potential and restricts the plants from taking up water and nutrients. The entry of high levels of Na+ ions into the roots of plants causes osmotic stress, which further disturbs the balance of essential nutrients within the plants resulting in ionic stress, and the superposition of these two stresses causes oxidative stress. The salinity stress causes major physiological process changes such as decreased plant growth, declined photosynthesis, and reduced yield.
Plants have developed regulatory pathways to acclimatize to salt stress. It comprises mainly signal perception, signal transduction, and responses to stress. The initial step for activating the salinity stress signaling cascade involves the recognition of stress through the plasma membrane receptors that sense physicochemical osmotic signals and chemical signals due to the excess Na+ and Cl− ions. (Figure 1). With the identification of the salinity stress, Na+ ions interact with the negatively charged components of the cell wall such as rhamnogalacturonan-II (RG-II) [4]. Moreover, with the increase in Na+ ions within the soil, several other carriers, or channels such as high-affinity K+ transporters (HKTs), glutamate receptors (GLRs), aquaporins, and nonselective cation channels (NSCCs) of root epidermal cells gets involved in the transport of Na+ ions into the plant cells (Figure 1). This changes the mechanical tension and turgor of the cell wall and leads to the opening of stretch-activated ion channels such as Ca2+ channels, causing an increase in the cytoplasmic Ca2+ ions that act as a secondary messenger of salt stress in addition to the ROS, diacylglycerol (DAG), inositol phosphates (IPs). Immediately, after the stress perception, the secondary messengers trigger a set of ROS-modulated protein kinases (PKs), protein phosphatases (PPs), and TFs that induce the stress-responsive genes for the plant’s adaptation to salinity stress.
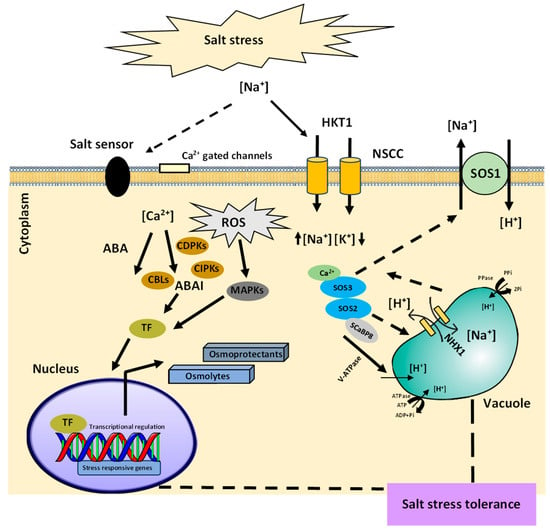
Figure 1. Schematic representation of plant salinity stress adaptation. Plant senses Na+ import intracellularly or extracellularly through both ABA-dependent and -independent signaling pathways. The regulatory pathways are induced through the initiation of various PK activities such as CDPKs, CIPKs, and CBLs. Ca2+ plays a major role in both the osmotic and ionic stress responses. The plants abate the ionic stress through either the exclusion of Na+ from the cells by SOS1 or through its sequestration into the vacuole with NHX1 transporters. The activation of transcriptional machinery increases or decreases certain stress-responsive gene expression that causes the synthesis of metabolites to counter the NaCl stress through the regulation of growth and metabolism.
The secondary messengers initiate the downstream responses to salinity stress and cause the modulation of stress-responsive genes through abscisic acid, ABA-dependent or -independent regulation mechanism (Figure 2). The common salt stress signal transduction pathways such as the salt overly sensitive (SOS) pathway and mitogen-activated protein kinase (MAPK) cascades play essential roles in the salt stress responses. The SOS pathway governs root ion homeostasis under salinity stress [6], whereas the high-osmolarity glycerol (HOG) MAPK cascade pathway controls the osmotic regulation during hyperosmotic stress [7]. In general, salt stress signal perception may activate multiple signaling pathways, which, along with crosstalk between the pathways, enable plants to adapt to salt stress.
The SOS signal transduction cascade that protect the cells from damage gets activated after the perception of a Ca2+ spike in the cytoplasm of root cells. SOS3 encodes a myristoylated calcium-binding protein, which acts as a primary calcium sensor to sense the increase in cytosolic Ca2+ triggered by excess of Na+ ions in the cytosol. Upon binding with Ca2+, SOS3 interacts with and activates the serine/threonine protein kinase SOS2 which is part of the SnRK3 protein kinases family (sucrose non-fermenting-1-related protein kinase-3). SOS3-like calcium-binding protein 8 (SCaBP8) has been displayed to be an alternative regulator of the activity of SOS2, which functions primarily in the Arabidopsis shoot, while SOS3 is more eminent in the roots [8]. SCaBP8 is phosphorylated by SOS2 which stabilizes the protein complex [9]. Phosphorylation of SOS3-like proteins by their interacting protein kinases is a common regulatory pathway for CBL/SCaBP–CIPK/PKS modules [10]. SOS3–SOS2 or SCaBP8–SOS2 interactions transfer SOS2 to the plasma membrane causing the activation of downstream target SOS1, a Na+/H+ antiporter. This results in the subsequent extrusion of toxic Na+ ions from the cytoplasm.
The PK, SOS, Ca2+, and certain phytohormones involved in this mechanism are governed by responsive genes, which are induced at the early and late stages of stress [11]. Early induced genes include TFs that are expressed swiftly after the perception of the stress, whereas the activation of late genes occurs gradually and may take several hours after the perception of stress to generate a response. However, these genes have sustained expression that modulates the required stress-responsive proteins. The products of late genes amplify the initial stress signal and trigger a secondary round of signaling, which may follow the same pathways as earlier or branch off into a new signaling pathway.
These pathways can include ROS detoxification which involves the antioxidative enzymes for scavenging the toxic free radicals. Salt sequestration into cell vacuoles by transporters (NHX1) is another core pathway used by plants to preserve a high cytosolic K+/Na+ ratio that regulates the toxic levels of salt ions in the cytosol for ion homeostasis (Figure 1). In addition, plants release and induce organic compounds, which are termed as osmolytes or compatible solutes [12], stress-responsive proteins [13], and osmoprotectants [14]. Possible outputs of these compounds include the expression of genes and activation of osmolyte biosynthesis enzymes. Most of the other changes induced by salinity stress can be considered to be involved in detoxification signaling. Furthermore, the osmolytes shield the essential proteins through the exclusion of hydrophilic molecules from their hydration sphere, which inhibits or reduces their interaction with water and protects their native structures. Moreover, the plants overcome the salt stress through cell wall modifications, membrane system adjustments, cell cycle, and cell division modifications.
3. Phytomorphological Adaptation of Plants in Saline Growth Conditions
Plants have developed various mechanisms in response to salinity stress that enables them to alter their morphological traits, resulting in sustained tolerance to high salt levels. The salt-tolerant plants possess shallow roots and develop many stilt or prop roots from their aerial branches of stem for efficient anchorage in saline soils. Plants grow these roots downward that enter the deep and tough strata of the soil. For instance, in Rhizophora mucronata, the stilt roots are strong and extensively developed, while in other species they are poorly developed such as in Rhizophora conjugate [15]. However, there are certain saline-tolerant plants with the absence of stilt roots. Furthermore, salt stress-tolerant plants consist of a large number of adventitious root buttresses that sprout from their basal parts, which provides them sufficient support in salt-affected soils. In the coastal areas, the saline soils are poorly aerated, containing a very small proportion of oxygen due to water logging. To overcome the lack of soil aeration, the plants develop negative geotropic roots, known as pneumatophores or breathing roots. The pneumatophores are developed from the underground roots and projected towards the air well above the surface of soil and water, which appears as peg-like structures with pointed tips. These roots surface possess numerous lenticels or pneumathodes and prominent aerenchyma that encloses the large air cavities internally. Lenticles are used for the gaseous exchanges in these roots and the aerenchyma is involved in the conduction of air down to the roots that are submerged. For example, in Bruguiera, the horizontal roots develop above the surface of the soil and then again bend down and enter deep into the soil forming knee-like structures [16]. The gaseous exchange is further facilitated by the pores in the aerial surface of the roots. However, in certain species such as Rhizophora, pneumatophores do not develop, due to which the respiratory activity under saline conditions is taken up by the upper aerial parts of descending stilt roots. Another key factor in coping with salinity stress is the plasticity of roots. The root morphological plasticity can restrict the buildup of salt ions in roots to allow the uptake of water under saline soils. It was found that at lower salt stress concentrations of 5 g L−1 NaCl, an abundance of root hairs was induced, but it gradually declined under greater salt concentrations of 10 g L−1 and 15 g L−1 NaCl in Bacopa monniera [17]. Moreover, in wheat plants under salinity stress, both the length and density of root per unit surface area were less than 25% and 40% in contrast to the hydroponically grown plant genotypes [18].
Stems of several salt-tolerant plants develop succulence, which is commonly found in Salicornia herbacia and Suaeda maritima. The succulence formation depends on the ratio of absorbed to free ions in the plant cells rather than the amount of absolute NaCl present. Succulence growth is stimulated with the increase in free salt ions above the threshold level within the plants. In salt-tolerant citrus species, Cleopatra mandarin, an increase in the succulence of leaves was found under the salinity stress [19]. According to Qi and Zhang, cell division gets inhibited during the salinity stress that promotes cell elongation [20]. This causes a decrease in the number of cells and an increase in the cell size, which commonly occurs in succulents. Fradera-Soler et al. (2022) stated that succulence is directly associated with the plant’s salt tolerance and the degree of their development can indicate the capability of plants to survive in highly saline habitats [21]. The temperate saline-tolerant plants are herbaceous, but the tropical ones are largely bushy and display dense cymose branching. Nevertheless, there are salt-tolerant plants such as the submerged marine angiosperms that do not become succulent.
The leaves in most salt-tolerant plants are generally smaller in size, thick, entire, and succulent, with a glassy appearance. In Cleopatra mandarin, salt-tolerant citrus species, an increase in the leaf thickness with a lower area/volume ratio of mesophyll cells was observed under salinity stress [19]. Similarly, under high salt stress conditions, increased leaf thickness was observed in Lawsonia inermis plants [22]. Furthermore, the coastal saline-tolerant plant leaves display an additional mode of adaptation to their environment. The leaves of these plants are densely covered with trichomes. In submerged marine plants, the leaves are thin with a poorly developed vascular system frequently having a green epidermis that is adapted to uptaking water and nutrients directly from the medium. Saline-tolerant plants develop lightweight fruits and seeds with fruit walls having several air chambers for fruit dispersal. In addition to these morphological adaptations, several salt-tolerant plants undergo physiological and biochemical adaptation mechanisms that are governed by a network of stress-responsive genes controlled by the TFs.
4. TFs Regulating Plant Responses to Salinity Stress
Several plant biochemical alterations during salinity stress occur due to the transcriptional changes that alter the plant’s growth and developmental processes. TFs are the core elements that control the whole-plant transcriptional responses as regulatory molecular switches of the genes [23]. These are instigated by ABA-dependent and -independent pathways that function either individually or through cross-linking with other TFs (Figure 2). They induce the expression of functional genes and initiate different salt tolerance phenomena in plants [24]. Based on the genome-wide analysis, various TFs belonging to different families, such as myeloblastosis (MYB), basic region/leucine zipper motif (bZIP), basic helix-loop-helix (bHLH), NAC, ethylene responsive factor (ERF/AP2), and WRKY have been studied for salt stress tolerance [25].
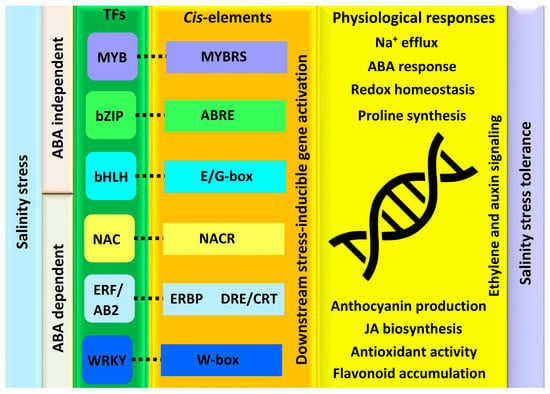
Figure 3. Overview of transcriptional modulation of salt stress in plants. Both the ABA-dependent and -independent pathway initiates the transcriptional regulatory networks. Salt stress signaling prompts transcription factors to bind to their corresponding cis-regulatory elements, which then triggers the expression of genes required for various physiological responses to cope with salinity stress.
5. Salinity Stress Tolerant Crop Wild Relatives (CWRs)
Contemporary agricultural systems heavily rely on a small proportion of highly productive crops. Only twenty plant species are cultivated to meet ninety percent of the world’s food calorie requirements. Among these, three crops such as wheat, rice, and maize are supplying about 60% of the total food [26]. More than 10,000 years ago, the domestication of these food crops occurred from their wild relatives that were distributed across a broad range of habitats, including salt marshes. The genetic traits largely contributed to the CWRs’ ability to thrive under salinity. During the course period, there was a significant transformation in the performance and genetic structure of the CWRs. By selectively breeding a small number of wild relatives with beneficial characteristics, such as non-brittle rachis, compact plant stature, and loss of germination inhibition, landraces with improved growth performance were created. However, in subsequent generations, this process led to a gradual reduction in genetic diversity. The shrinkage in the genetic diversity of crops has been further intensified by contemporary techniques of breeding plants, which focus on producing improved yielding cultivars by crossing landraces that are productive and ignoring the wild relatives that possess larger genetic diversity but have poor agronomic characteristics. Such narrowing of genetic diversity has been experimentally confirmed [27]. Moreover, the shift of farmers from cultivating local crop varieties and landraces to genetically uniform and high-yielding varieties has led to a loss of about 75% of genetic diversity in crops [26].
Importantly, as present techniques of plant breeding tend to be performed in optimized agricultural settings, the genetic elements of abiotic stress tolerance including salinity, are frequently amongst the lost fractions. Hence, the attempts to find genes that provide tolerance to the salinity stress within the commercially used varieties have produced only restrictive outcomes. Therefore, the CWRs are essential resources for genetic elements of salinity stress tolerance that can be utilized in contemporary agricultural breeding [28]. For example, Hordeum marinum, sea barley grass is classified as a wild halophyte, whereas other species of Hordeum are considered as glycophytes. A physiological analysis revealed that sea barley grass (H559) demonstrated greater salinity tolerance compared to the barley genotypes XZ113 and Golden Promise [29]. Another study found that certain accessions of Tibetan wild barley exhibited superior salinity tolerance in contrast to the well-established saline-tolerant barley cultivar CM72 [30]. Oryza coarctata (Porteresia coarctata) is a wild halophytic rice found in the coastal environments, which possessed significant tolerance to both the salinity stress and submergence [31]. Similarly, several CWRs were identified to be salt stress tolerant (Table 1).
Table 1. Salt tolerant wild relatives of various crop species.
| Wild Species | Plants | Salt Stress Tolerance QTLs/TFs/Genes | Source |
|---|---|---|---|
| Gossypium tomentosum | Cotton | qRL-Chr16-1 | [32] |
| Gossypium davidsonii | KUP1, KUP2, KUP11, SKOR, NCED3, PYR/PYL/RCARs, SnRK2s, AtMYB20, and PP2C | [33] | |
| Gossypium aridum | GarWRKY17 and GarWRKY104 | [34] | |
| Solanum hirsutum | Tomato | aox-c6.1, fla6.1, cat-c12.1, pox-s7.1, pox-s12.1, phe9.1, phe11.1, phe-c2.2, phe-c8.1 | [35] |
| Solanum parviflorum | DREB1A and Vp1.1 | [36] | |
| Arachis diogoi | Groundnut | AdDjSKI | [37] |
| Arachis duranensis | AdNACs | [38] | |
| Arachis glabrata | MYB, AP2, GRAS, bHLH, C3H, WRKY, C2H2 and ARF | [39] | |
| Ipomoea imperati | Potato | AP2/EREBP, bHLH, HD-ZIP and MYB | [40] |
| Oryza rufipogon | Rice | qST1-1, qST5-1, qST5-2, qST9, qST10, qST11-1, qST11-2, qST12, qST1-1, qST1-2, qST7, qST9, qST10, qST11-1, qST11-2, qST12 | [41] |
| Tripsacum dactyloides | Maize | CH3, MYB, HB, SNF2, AUX, and SET | [42] |
| Glycine soja | Soybean | Ncl2 | [43] |
| Medicago ruthenica | Alfalfa | NAC, C2H2, and CAMTA | [44] |
The broad variation in the salt stress tolerance quantitative trait locus (QTL), TFs, and genes of CWRs can serve as valuable sources for the enhancement of the cultivated species salinity stress tolerance. However, this genetic diversity needs to be examined further for the identification of essential molecular pathways and the utilization of various elements involved in them.
This entry is adapted from the peer-reviewed paper 10.3390/ijms24129813
References
- Qadir, M.; Quillerou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Res. Forum. 2014, 38, 282–295.
- FAO. The Future of Food and Agriculture—Trends and Challenges. 2017. Available online: https://agrinatura-eu.eu/news/the-future-of-food-and-agriculture-trends-and-challenges/
- Kapazoglou, A.; Gerakari, M.; Lazaridi, E.; Kleftogianni, K.; Sarri, E.; Tani, E.; Bebeli, P.J. Crop wild relatives: A valuable source of tolerance to various abiotic stresses. Plants 2023, 12, 328.
- Quezada-Martinez, D.; Addo Nyarko, C.P.; Schiessl, S.V.; Mason, A.S. Using wild relatives and related species to build climate resilience in Brassica crops. Theor. Appl. Genet. 2021, 134, 1711–1728.
- Kashyap, A.; Garg, P.; Tanwar, K.; Sharma, J.; Gupta, N.C.; Ha, P.T.T.; Bhattacharya, R.C.; Mason, A.S.; Rao, M. Strategies for utilization of crop wild relatives in plant breeding programs. Theor. Appl. Genet. 2022, 135, 4151–4167.
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) pathway: Established and emerging roles. Mol. Plant 2013, 6, 275–286.
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78.
- Quan, R.; Lin, H.; Mendoza, I.; Zhang, Y.; Cao, W.; Yang, Y.; Shang, M.; Chen, S.; Pardo, J.M.; Guo, Y. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 2007, 19, 1415–1431.
- Lin, H.; Yang, Y.; Quan, R.; Mendoza, I.; Wu, Y.; Du, W.; Zhao, S.; Schumaker, K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 2009, 21, 1607–1619.
- Du, W.; Lin, H.; Chen, S.; Wu, Y.; Zhang, J.; Fuglsang, A.T.; Palmgren, M.G.; Wu, W.; Guo, Y. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 2011, 156, 2235–2243.
- Shah, W.H.; Rasool, A.; Saleem, S.; Mushtaq, N.U.; Tahir, I.; Hakeem, K.R.; Rehman, R.U. Understanding the integrated pathways and mechanisms of transporters, protein kinases, and transcription factors in plants under salt stress. Int. J. Genom. 2021, 2021, 5578727.
- Das, A.B.; Strasser, R.J. Salinity-induced genes and molecular basis of salt-tolerant strategies in Mangroves. In Molecular Stress Physiology of Plants; Rout, G.R., Das, A.B., Eds.; Springer: Cham, Switzerland, 2013; pp. 53–86.
- Abdul Aziz, M.; Sabeem, M.; Mullath, S.K.; Brini, F.; Masmoudi, K. Plant group II LEA proteins: Intrinsically disordered structure for multiple functions in response to environmental stresses. Biomolecules 2021, 11, 1662.
- Kronzucker, H.J.; Britto, D.T. Sodium transport in plants: A critical review. New Phytol. 2011, 189, 54–81.
- Shahari, R.; Shamin-Shazwan, K.; Amri, C.; Kassim, Z.; Ahmad, Z. Morphological structures of Rhizophora apiculata blume. and Rhizophora mucronata lam. Sci. Herit. J. 2020, 5, 1–4.
- Basyuni, M.; Nainggolan, S.S.; Qurrahman, T.; Hasibuan, P.A.Z.; Sumaiyah, S.; Sumardi, S.; Siregar, E.S.; Nuryawan, A. Effect of salt and fresh water concentration on polyisoprenoid content in Bruguiera cylindrica seedlings. Open Access Maced. J. Med. Sci. 2019, 7, 3803–3806.
- Ali, G.; Srivastava, P.S.; Iqbal, M. Structural changes in root and shoot of Bacopa monniera in response to salt stress. J. Plant Biol. 1999, 42, 222.
- Robin, A.H.K.; Matthew, C.; Uddin, M.J.; Bayazid, K.N. Salinity-induced reduction in root surface area and changes in major root and shoot traits at the phytomere level in wheat. J. Exp. Bot. 2016, 67, 3719–3729.
- Romero-Aranda, R.; Moya, J.L.; Tadeo, F.R.; Legaz, F.; Primo-Millo, E.; Talon, M. Physiological and anatomical disturbances induced by chloride salts in sensitive and tolerant citrus: Beneficial and detrimental effects of cations. Plant Cell Environ. 1998, 21, 1243–1253.
- Qi, F.; Zhang, F. Cell cycle regulation in the plant response to stress. Front. Plant Sci. 2020, 10, 1765.
- Fradera-Soler, M.; Grace, O.M.; Jørgensen, B.; Mravec, J. Elastic and collapsible: Current understanding of cell walls in succulent plants. J. Exp. Bot. 2022, 73, 2290–2307.
- Fernández-García, N.; Olmos, E.; Bardisi, E.; García-De la Garma, J.; López-Berenguer, C.; Rubio-Asensio, J.S. Intrinsic water use efficiency controls the adaptation to high salinity in a semi-arid adapted plant, henna (Lawsonia inermis L.). J. Plant Physiol. 2014, 171, 64–75.
- Zhang, X.; Cheng, Z.; Fan, G.; Yao, W.; Li, W.; Chen, S.; Jiang, T. Functional analysis of PagNAC045 transcription factor that improves salt and ABA tolerance in transgenic tobacco. BMC Plant Biol. 2022, 22, 261.
- Khoso, M.A.; Hussain, A.; Ritonga, F.N.; Ali, Q.; Channa, M.M.; Alshegaihi, R.M.; Meng, Q.; Ali, M.; Zaman, W.; Brohi, R.D.; et al. WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 2022, 8, 1039329.
- Gao, F.; Zhou, J.; Deng, R.Y.; Zhao, H.X.; Li, C.L.; Chen, H.; Suzuki, T.; Park, S.U.; Wu, Q. Overexpression of a tartary buckwheat R2R3-MYB transcription factor gene, FtMYB9, enhances tolerance to drought and salt stresses in transgenic Arabidopsis. J. Plant Physiol. 2017, 214, 81–90.
- Buchanan-Wollaston, V.; Wilson, Z.; Tardieu, F.; Beynon, J.; Denby, K. Harnessing diversity from ecosystems to crops to genes. Food Energy Secur. 2017, 6, 19–25.
- Abbo, S.; Pinhasi Van-Oss, R.; Gopher, A.; Saranga, Y.; Ofner, I.; Peleg, Z. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014, 19, 351–360.
- Isayenkov, S.V. Genetic sources for the development of salt tolerance in crops. Plant Growth Regul. 2019, 89, 1–17.
- Huang, L.; Kuang, L.H.; Li, X.; Wu, L.; Wu, D.; Zhang, G. Metabolomic and transcriptomic analyses reveal the reasons why Hordeum marinum has higher salt tolerance than Hordeum vulgare. Environ. Exp. Bot. 2018, 156, 48–61.
- Qiu, L.; Wu, D.; Ali, S.; Cai, S.; Dai, F.; Jin, X.; Wu, F.; Zhang, G. Evaluation of salinity tolerance and analysis of allelic function of HvHKT1 and HvHKT2 in Tibetan wild barley. Theor. Appl. Genet. 2011, 122, 695–703.
- Garg, R.; Verma, M.; Agrawal, S.; Shankar, R.; Majee, M.; Jain, M. Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res. 2014, 21, 69–84.
- Oluoch, G.; Zheng, J.; Wang, X.; Khan, M.K.R.; Zhou, Z.; Cai, X.; Wang, C.; Wang, Y.; Li, X.; Wang, H.; et al. QTL mapping for salt tolerance at seedling stage in the interspecific cross of Gossypium tomentosum with Gossypium hirsutum. Euphytica 2016, 209, 223–235.
- Zhang, F.; Zhu, G.; Du, L.; Shang, X.; Cheng, C.; Yang, B.; Hu, Y.; Cai, C.; Guo, W. Genetic regulation of salt stress tolerance revealed by RNA-Seq in cotton diploid wild species, Gossypium davidsonii. Sci. Rep. 2016, 6, 20582.
- Fan, X.; Guo, Q.; Xu, P.; Gong, Y.; Shu, H.; Yang, Y.; Ni, W.; Zhang, X.; Shen, X. Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PLoS ONE 2015, 10, e0126148.
- Frary, A.; Göl, D.; Keleş, D.; Ökmen, B.; Pınar, H.; Şığva, H.Ö.; Yemenicioğlu, A.; Doğanlar, S. Salt tolerance in Solanum pennellii: Antioxidant response and related QTL. BMC Plant Biol. 2010, 10, 58.
- Rao, E.S.; Kadirvel, P.; Symonds, R.C.; Geethanjali, S.; Thontadarya, R.N.; Ebert, A.W. Variations in DREB1A and VP1.1 genes show association with salt tolerance traits in wild tomato (Solanum pimpinellifolium). PLoS ONE 2015, 10, e0132535.
- Rampuria, S.; Bag, P.; Rogan, C.J.; Sharma, A.; Gassmann, W.; Kirti, P.B. Pathogen-induced AdDjSKI of the wild peanut, Arachis diogoi, potentiates tolerance of multiple stresses in E. coli and tobacco. Plant Sci. Int. J. Exp. Plant Biol. 2018, 272, 62–74.
- Yuan, C.; Li, C.; Lu, X.; Zhao, X.; Yan, C.; Wang, J.; Sun, Q.; Shan, S. Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol. 2020, 20, 454.
- Zhao, C.; He, L.; Xia, H.; Zhou, X.; Geng, Y.; Hou, L.; Li, P.; Li, G.; Zhao, S.; Ma, C.; et al. De novo full length transcriptome analysis of Arachis glabrata provides insights into gene expression dynamics in response to biotic and abiotic stresses. Genomics 2021, 113, 1579–1588.
- Zhao, C.; He, L.; Xia, H.; Zhou, X.; Geng, Y.; Hou, L.; Li, P.; Li, G.; Zhao, S.; Ma, C.; et al. De novo full length transcriptome analysis of Arachis glabrata provides insights into gene expression dynamics in response to biotic and abiotic stresses. Genomics 2021, 113, 1579–1588.
- Wang, S.; Cao, M.; Ma, X.; Chen, W.; Zhao, J.; Sun, C.; Tan, L.; Liu, F. Integrated RNA sequencing and QTL mapping to identify candidate genes from Oryza rufipogon associated with salt tolerance at the seedling stage. Front. Plant Sci. 2017, 8, 1427.
- Li, X.; Wang, X.; Ma, Q.; Zhong, Y.; Zhang, Y.; Zhang, P.; Li, Y.; He, R.; Zhou, Y.; Li, Y.; et al. Integrated single-molecule real-time sequencing and RNA sequencing reveal the molecular mechanisms of salt tolerance in a novel synthesized polyploid genetic bridge between maize and its wild relatives. BMC Genom. 2023, 24, 55.
- Lee, J.D.; Shannon, J.G.; Vuong, T.D.; Nguyen, H.T. Inheritance of salt tolerance in wild soybean (Glycine soja Sieb. and Zucc.) accession PI483463. J. Hered. 2009, 100, 798–801.
- Yin, M.; Zhang, S.; Du, X.; Mateo, R.G.; Guo, W.; Li, A.; Wang, Z.; Wu, S.; Chen, J.; Liu, J.; et al. Genomic analysis of Medicago ruthenica provides insights into its tolerance to abiotic stress and demographic history. Mol. Ecol. Resour. 2021, 21, 1641–1657.
This entry is offline, you can click here to edit this entry!
