Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Hematology
Major advances in the understanding of acute myeloid leukemia (AML) pathogenesis, together with technological progress, have led us into a new era in the diagnosis and follow-up of patients with AML. A combination of immunophenotyping, cytogenetic and molecular studies are required for AML diagnosis, including the use of next-generation sequencing (NGS) gene panels to screen all genetic alterations with diagnostic, prognostic and/or therapeutic value.
- AML
- diagnosis
- flow cytometry
1. Introduction
Acute myeloid leukemia (AML) is characterized by a high biological heterogeneity both at diagnosis and during disease evolution. An exhaustive characterization of immunophenotypic and molecular profiles is required to guide the clinical management of AML patients. In recent years, growing evidence has shown the prominent role of cytogenetic and mutational data in AML and, accordingly, the recently updated World Health Organization (WHO) [1] and International Consensus Classification (ICC) [2] classifications of myeloid neoplasms as well as the European LeukemiaNet (ELN) recommendations [3] have incorporated genetics into their diagnostic, prognostic and therapeutic algorithms. In this scenario, a combination of immunophenotyping and genetic studies are required for AML diagnosis and follow-up. Measurable residual disease (MRD) has strong prognostic and predictive value in AML, and its accurate monitoring is crucial for disease management. The plethora of aberrant phenotypic and molecular characteristics of AML cells represents a chance to track this disease but, at the same time, constitutes a major challenge for hematopathology laboratories.
2. Diagnosis
2.1. Flow Cytometry for Lineage Assessment
Multiparametric flow cytometry (MFC) consists of the recognition of cells based on antigen detection through a combination of fluorochrome-labeled monoclonal antibodies, which allows phenotypic characterization and the quantification of a specific cell population.
When acute leukemia is suspected, morphologic and immunophenotypic examinations of the bone marrow and peripheral blood are the quickest techniques to confirm the diagnosis (Figure 1). The latter is also essential when assigning the leukemic lineage: a crucial step that determines further diagnostic tests and therapeutic strategies. Lineage-assigning antigens are shown in Table 1. Myeloperoxidase (MPO) expression is the hallmark of myeloid commitment, though it is not always present. AML with minimal differentiation is defined by an absence of lineage-assigning antigens in combination with the expression of at least two other myeloid-related antigens (e.g., CD13, CD33, CD117). Monocytic AML often loses MPO expression and is characterized by specific markers such as CD11c, CD14, CD64 and lysozyme.
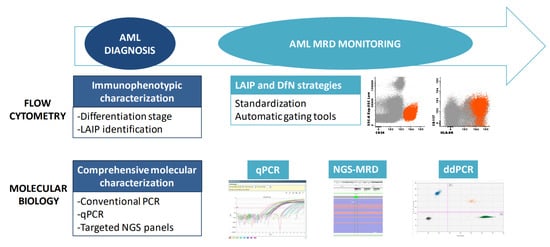
Figure 1. Tools for diagnosis and monitoring of AML. A combination of immunophenotyping and molecular studies is required for AML diagnosis and MRD monitoring. MRD: Measurable residual disease; LAIP: Leukemia-associated immunophenotype; DfN: Different from normal; qPCR: quantitative PCR; ddPCR: Droplet digital PCR.
Table 1. Flow cytometry diagnostic and MRD markers.
| Diagnostic Markers | |
|---|---|
| Lineage assigning antigens | |
| Myeloperoxidase | Myeloid lineage |
| CD11c, CD14, CD64, lysozyme | Myeloid lineage with monocytic differentiation |
| CD19 | B-lineage. Requires also at least one (CD19 strong) or two (CD19 weak) from CD22, CD10 and CD79a |
| CD3 (surface or cytoplasmic) | T-lineage |
| Myeloid differentiation-associated antigens | |
| CD13, CD33, CD11b, CD15, CD64 | Myeloid |
| CD14, CD36, CD64, CD4, CD11c | Monocytic |
| CD41, CD42b, CD61, CD36 | Megakaryocytic |
| CD235a, CD71 strong, CD105, CD36 | Erythroid |
| CD203c, CD123 | Basophil |
| CD123, CD4, HLA-DR strong, CD303, CD304 | Dendritic |
| CD117 strong | Mastocytic |
| MRD markers | |
| Basic markers | |
| CD34, CD117, HLA-DR, CD45, CD13, CD33 | Myeloid precursor identification |
| CD7, CD56 | Lymphoid antigen aberrancies |
| Other useful markers | |
| CD64, CD14, CD11b, CD4 | Monocytic |
| CD19, CD2, CD5 | Lymphoid antigen aberrancies |
| CD38, CD123, CD133 | Leukemia stem cell identification |
AML differentiation, maturation stage and the aberrant co-expression of lymphoid markers can be only addressed with a broad panel of antibodies. Morphologic and immunophenotypic criteria for cases without recurrent genetic abnormalities have been updated in the latest WHO classification [1]. Even when this information lacks its independent prognostic impact, it often correlates with particular genetic lesions. For example, acute promyelocytic leukemia (APL) exhibits a characteristic immunophenotype (CD34−, HLA-DR−, CD117+, CD15−) that can prompt PML::RARA gene fusion evaluation and consequent management until the molecular result is available. Another example is the association between the co-expression of CD25, CD123 and CD99 on myeloid blast and FLT3 mutations [4].
The Euroflow Consortium has developed, through serial testing, an acute leukemia orientation tube (ALOT), which includes most of the lineage-defining markers, and a panel for AML that covers the different myeloid lineages and most frequent lymphoid aberrancies [5]. Moreover, the standardization of MFC and the building of reference databases with normal and acute leukemia samples allows for automated gating identification of leukocyte subpopulations and classification of blast cells [6,7].
The use of a comprehensive panel of antibodies also allows for the recognition of leukemia-associated immunophenotype (LAIP) patterns that can be of interest during follow-up. Finally, new therapies targeting the surface antigens of myeloid blasts are under development. In the future, flow cytometry could be useful when identifying potential candidates for treatment against not-universally expressed antigens, as could be chimeric antigen receptor therapy against CD123 [8].
2.2. Molecular Biology
Over the last decade, genomic studies based on Next-Generation Sequencing (NGS) have dissected the molecular profile of AML, with the description of new mutations, copy number variations and recurrent fusion genes [9,10]. The growth in molecular knowledge has prompted the update of both diagnostic and management recommendations for AML patients. The recently updated WHO, ICC and ELN classifications have undertaken a major role in genetic data and have been integrated into diagnostic and prognostic algorithms (Table 2) [1,2,3]. Genetic analyses currently mandatory in the evaluation of AML are conventional cytogenetics together with the screening of fusion genes and of gene mutations, whose number has significantly increased in comparison with prior classifications (Figure 1). Recommendations for molecular assessments in AML include the following genes: FLT3 (both internal tandem duplications, ITD, and tyrosine kinase domain, TKD, mutations), IDH1 and IDH2 as therapeutic targets, and NPM1, CEBPA, DDX41, TP53, ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1 and ZRSR2 as diagnostic/prognostic disease markers. Furthermore, the screening of the most prevalent gene rearrangements include PML::RARA, RUNX1::RUNX1T1, CBFB::MYH11, KMT2A rearrangements, BCR::ABL1 and others and should be performed when rapid information is needed for the clinical management of a patient, or if the morphology and/or immunophenotype are highly suggestive of the presence of a particular fusion gene [3].
Table 2. Classification of AML according to ICC and WHO 2022.
| ICC 2022 | WHO 2022 | ||
|---|---|---|---|
| Category | Blasts Required for Diagnosis | Category | Blasts Required for Diagnosis |
| AML with Recurrent Genetic Abnormalities | AML with Defining Genetic Abnormalities | ||
| Acute promyelocytic leukemia (APL) with t(15;17)(q24.1;q21.2)/PML::RARA | ≥10% | Acute promyelocytic leukemia (APL) with PML::RARA fusion | No threshold |
| APL with other RARA rearrangements | ≥10% | ||
| AML with t(8;21)(q22;q22.1)/RUNX1::RUNX1T1 | ≥10% | AML with RUNX1::RUNX1T1 fusion | No threshold |
| AML with inv(16)(p13.1q22) or t(16;16)(p13.1;q22)/CBFB::MYH11 | ≥10% | AML with CBFB::MYH11 fusion | No threshold |
| AML with t(9;11)(p21.3;q23.3)/MLLT3::KMT2A | ≥10% | AML with KMT2A rearrangement | No threshold |
| AML with other KMT2A rearrangements | ≥10% | ||
| AML with t(6;9)(p22.3;q34.1)/DEK::NUP214 | ≥10% | AML with DEK::NUP214 fusion | No threshold |
| AML with inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2)/GATA2; MECOM(EVI1) |
≥10% | AML with MECOM rearrangement | No threshold |
| AML with other MECOM rearrangements | ≥10% | ||
| AML with other rare recurring translocations | ≥10% | AML with NUP98 rearrangement | No threshold |
| AML with RBM15::MRTFA fusion | No threshold | ||
| AML with t(9;22)(q34.1;q11.2)/BCR::ABL1 | ≥20% | AML with BCR::ABL1 fusion | ≥20% |
| AML with mutated NPM1 | ≥10% | AML with NPM1 mutation | No threshold |
| AML with in-frame bZIP CEBPA mutations | ≥10% | AML with CEBPA mutation | ≥20% |
| AML and MDS/AML with mutated TP53 | 10–19% (MDS/AML) and ≥20% (AML) |
||
| AML and MDS/AML with myelodysplasia-related gene mutations Defined by mutations in ASXL1, BCOR, EZH2, RUNX1, SF3B1, SRSF2, STAG2, U2AF1, or ZRSR2 |
10–19% (MDS/AML) and ≥20% (AML) |
AML, myelodysplasia-related Defined by one or more cytogenetics of molecular alterations:
|
≥20% |
| AML with myelodysplasia-related cytogenetic abnormalities Defined by a complex karyotype (≥3 unrelated clonal chromosomal abnormalities in the absence of other class-defining recurring genetic abnormalities), del(5q)/t(5q)/add(5q), −7/del(7q), +8, del(12p)/t(12p)/add(12p), i(17q),−17/add(17p) or del(17p), del(20q), and/or idic(X)(q13) clonal abnormalities |
10–19% (MDS/AML) and ≥20% (AML) |
||
| AML with other defined genetic alterations | No threshold | ||
| AML without recurrent genetic abnormalities | AML without defining genetic abnormalities | ||
| AML not otherwise specified (NOS) | 10–19% (MDS/AML) and ≥20% (AML) |
AML defined by differentiation | ≥20% |
In this scenario, the screening of single genes is progressively being replaced by targeted NGS panels, which, in addition, could offer a combined solution for the parallel analysis of DNA for mutations and RNA for fusions in the same assay. However, the main limitation of NGS technology is that it requires batching to be cost-effective, making it unrealistic for most diagnostic laboratories to report NGS results within a week. In this context, conventional PCR-based techniques are still being used for the screening of relevant markers that require a rapid turn-around time (3–5 days), including NPM1, FLT3, IDH1/2, and fusion genes. In particular, FLT3-ITD evaluation is recommended to be performed by capillary electrophoresis [11] because it cannot be performed robustly by NGS.
Among the most relevant molecular changes included in the new classifications is the description of some new AML types with fusion genes such as DEK::NUP214, BCR::ABL1 or rearrangements involving KMT2A, MECOM and NUP98 (Table 2). Another remarkable change refers to the recognition of particular clinicobiological entities associated with adverse risk, AML with a TP53 mutation and the renamed AML with myelodysplasia-related gene mutations (ICC 2022)/AML myelodysplasia-related (WHO 2022), which harbor specific cytogenetic and molecular alterations beyond those previously considered ASXL1 and RUNX1 (Table 2). Regarding AML with CEBPA mutations, it remains a favorable prognostic entity, but a biallelic mutation is not required, according to several studies published; a single in-frame mutation in the basic leucine zipper (bZIP) region of CEBPA is sufficient for diagnosis [3,12,13]. At the prognostic level, all AML patients with FLT3-ITD are classified in the intermediate risk group irrespective of the FLT3-ITD ratio and the presence of the NPM1 mutation. Furthermore, the presence of adverse-risk cytogenetic aberrations in NPM1-mutated AML is now also indicative of high risk [3].
The study of an additional set of genes, including ANKRD26, BCORL1, BRAF, CBL, CSF3R, DNMT3A, ETV6, GATA2, JAK2, KIT, KRAS, NRAS, NF1, PHF6, PPM1D, PTPN11, RAD21, SETBP1, TET2 and WT1, is recommended at AML diagnosis, and some of them can also be used for MRD monitoring [3,14]. When AML with germline predisposition is suspected, mutational studies should be complemented with an extended NGS panel covering known predisposing genes together with genetic counseling. Both ICC and WHO updated classifications include specific categories of myeloid neoplasms with a germline predisposition characterized by the presence of germline inherited mutations and specific clinical features [1,2].
This entry is adapted from the peer-reviewed paper 10.3390/curroncol30060395
This entry is offline, you can click here to edit this entry!
