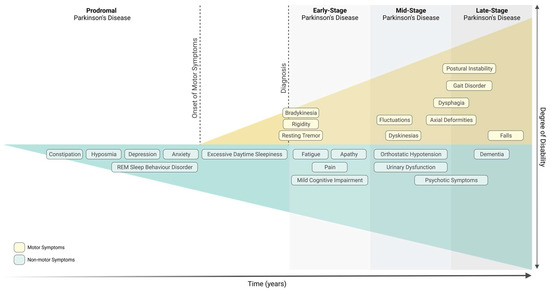
Parkinson’s Disease (PD), the second most common neurodegenerative disorder, is characterised by the severe loss of dopaminergic neurons in the Substantia Nigra pars compacta (SNpc) and by the presence of Lewy bodies. PD is diagnosed upon the onset of motor symptoms, such as bradykinesia, resting tremor, rigidity, and postural instability. It is accepted that motor symptoms are preceded by non-motor features, such as gastrointestinal dysfunction. In fact, it has been proposed that PD might start in the gut and spread to the central nervous system. Growing evidence reports that the gut microbiota, which has been found to be altered in PD patients, influences the function of the central and enteric nervous systems. Altered expression of microRNAs (miRNAs) in PD patients has also been reported, many of which regulate key pathological mechanisms involved in PD pathogenesis, such as mitochondrial dysfunction and immunity. It remains unknown how gut microbiota regulates brain function, however miRNAs have been highlighted as important players. Remarkably, numerous studies have depicted the ability of miRNAs to modulate and be regulated by the host’s gut microbiota.
- microRNAs
- Parkinson’s disease
- gut microbiota
1. Introduction
Parkinson’s Disease
2. miRNA Involvement in Parkinson’s Disease
2.1. Human and Animal Studies
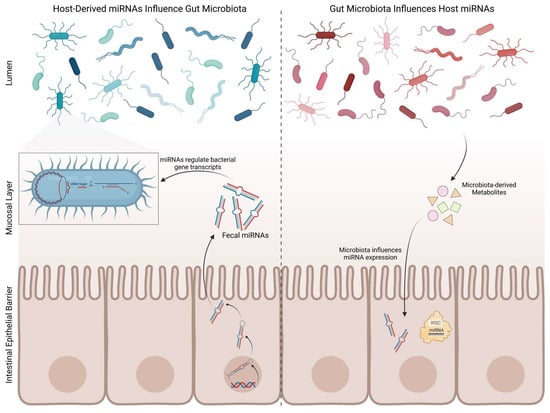
2.2. Gut Microbiota and microRNAs
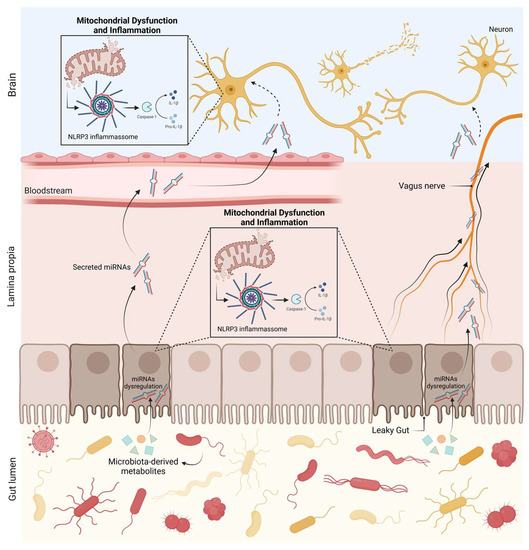
3. Conclusions and Future Perspectives
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines11051349
References
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. https://doi.org/10.1016/s0140-6736(21)00218-x
- Jankovic, J.; Tan, E.K. Parkinson’s disease: Etiopathogenesis and treatment. J. Neurol. Neurosurg. Psychiatry 2020, 91, 795–808. https://doi.org/10.1136/jnnp-2019-322338
- Mahlknecht, P.; Marini, K.; Werkmann, M.; Poewe, W.; Seppi, K. Prodromal Parkinson’s disease: Hype or hope for disease-modification trials? Transl. Neurodegener. 2022, 11, 11. https://doi.org/10.1186/s40035-022-00286-1
- Cuenca, L.; Gil-Martinez, A.L.; Cano-Fernandez, L.; Sanchez-Rodrigo, C.; Estrada, C.; Fernandez-Villalba, E.; Herrero Ezquerro, M.T. Parkinson’s disease: A short story of 200 years. Histol. Histopathol. 2019, 34, 573–591. https://doi.org/10.14670/HH-18-073
- Braak, H.; Del Tredici, K.; Rüb, U.; de Vos, R.A.; Steur, E.N.J.; Braak, E. Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 2003, 24, 197–211. https://doi.org/10.1016/s0197-4580(02)00065-9
- Rietdijk, C.D.; Perez-Pardo, P.; Garssen, J.; Van Wezel, R.J.A.; Kraneveld, A.D. Exploring Braak’s Hypothesis of Parkinson’s Disease. Front. Neurol. 2017, 8, 37. https://doi.org/10.3389/fneur.2017.00037
- Chen, H.; Wang, K.; Scheperjans, F.; Killinger, B. Environmental triggers of Parkinson’s disease—Implications of the Braak and dual-hit hypotheses. Neurobiol. Dis. 2022, 163, 105601. https://doi.org/10.1016/j.nbd.2021.105601
- Simon, D.K.; Tanner, C.M.; Brundin, P. Parkinson Disease Epidemiology, Pathology, Genetics, and Pathophysiology. Clin. Geriatr. Med. 2019, 36, 1–12. https://doi.org/10.1016/j.cger.2019.08.002
- Borghammer, P.; Horsager, J.; Andersen, K.; Berge, N.V.D.; Raunio, A.; Murayama, S.; Parkkinen, L.; Myllykangas, L. Neuropathological evidence of body-first vs. brain-first Lewy body disease. Neurobiol. Dis. 2021, 161, 105557. https://doi.org/10.1016/j.nbd.2021.105557
- Nuzum, N.D.; Loughman, A.; Szymlek-Gay, E.A.; Teo, W.-P.; Hendy, A.M.; Macpherson, H. To the Gut Microbiome and Beyond: The Brain-First or Body-First Hypothesis in Parkinson’s Disease. Front. Microbiol. 2022, 13, 791213. https://doi.org/10.3389/fmicb.2022.791213
- Pajares, M.; Rojo, A.I.; Manda, G.; Boscá, L.; Cuadrado, A. Inflammation in Parkinson’s Disease: Mechanisms and Therapeutic Implications. Cells 2020, 9, 1687. https://doi.org/10.3390/cells9071687
- Belvisi, D.; Pellicciari, R.; Fabbrini, A.; Costanzo, M.; Ressa, G.; Pietracupa, S.; De Lucia, M.; Modugno, N.; Magrinelli, F.; Dallocchio, C.; et al. Relationship between risk and protective factors and clinical features of Parkinson’s disease. Park. Relat. Disord. 2022, 98, 80–85. https://doi.org/10.1016/j.parkreldis.2022.04.017
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; et al. Mutation in the α-Synuclein Gene Identified in Families with Parkinson’s Disease. Science 1997, 276, 2045–2047. https://doi.org/10.1126/science.276.5321.2045
- Jia, F.; Fellner, A.; Kumar, K.R. Monogenic Parkinson’s Disease: Genotype, Phenotype, Pathophysiology, and Genetic Testing. Genes 2022, 13, 471. https://doi.org/10.3390/genes13030471
- Guedes, L.C.; Mestre, T.; Outeiro, T.F.; Ferreira, J.J. Are genetic and idiopathic forms of Parkinson’s disease the same disease? J. Neurochem. 2020, 152, 515–522. https://doi.org/10.1111/jnc.14902
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Prim. 2017, 3, 17013. https://doi.org/10.1038/nrdp.2017.13
- Rodriguez, M.; Rodriguez-Sabate, C.; Morales, I.; Sanchez, A.; Sabate, M. Parkinson’s disease as a result of aging. Aging Cell 2015, 14, 293–308. https://doi.org/10.1111/acel.12312
- Parkinson, J. An Essay on the Shaking Palsy. J. Neuropsychiatry Clin. Neurosci. 1817, 14, 223–236, discussion 222. https://doi.org/10.1176/jnp.14.2.223
- Yao, S. MicroRNA biogenesis and their functions in regulating stem cell potency and differentiation. Biol. Proced. Online 2016, 18, 8. https://doi.org/10.1186/s12575-016-0037-y
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. https://doi.org/10.3389/fendo.2018.00402
- Nies, Y.H.; Najib, N.H.M.; Lim, W.L.; Kamaruzzaman, M.A.; Yahaya, M.F.; Teoh, S.L. MicroRNA Dysregulation in Parkinson’s Disease: A Narrative Review. Front. Neurosci. 2021, 15, 660379. https://doi.org/10.3389/fnins.2021.660379
- Schulz, J.; Takousis, P.; Wohlers, I.; Itua, I.O.; Dobricic, V.; Rücker, G.; Binder, H.; Middleton, L.; Ioannidis, J.P.; Perneczky, R.; et al. Meta-analyses identify differentially expressed microRNAs in Parkinson’s disease. Ann. Neurol. 2019, 85, 835–851. https://doi.org/10.1002/ana.25490
- Hoss, A.G.; Labadorf, A.; Beach, T.G.; Latourelle, J.C.; Myers, R.H. microRNA Profiles in Parkinson’s Disease Prefrontal Cortex. Front. Aging Neurosci. 2016, 8, 36. https://doi.org/10.3389/fnagi.2016.00036
- Cho, H.J.; Liu, G.; Jin, S.M.; Parisiadou, L.; Xie, C.; Yu, J.; Sun, L.; Ma, B.; Ding, J.; Vancraenenbroeck, R.; et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2013, 22, 608–620. https://doi.org/10.1093/hmg/dds470
- Briggs, C.E.; Wang, Y.; Kong, B.; Woo, T.-U.W.; Iyer, L.K.; Sonntag, K.C. Midbrain dopamine neurons in Parkinson׳s disease exhibit a dysregulated miRNA and target-gene network. Brain Res. 2015, 1618, 111–121. https://doi.org/10.1016/j.brainres.2015.05.021
- Tatura, R.; Kraus, T.; Giese, A.; Arzberger, T.; Buchholz, M.; Höglinger, G.; Müller, U. Parkinson’s disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Park. Relat. Disord. 2016, 33, 115–121. https://doi.org/10.1016/j.parkreldis.2016.09.028
- Cardo, L.F.; Coto, E.; Ribacoba, R.; Menéndez, M.; Moris, G.; Suárez, E.; Alvarez, V. MiRNA Profile in the Substantia Nigra of Parkinson’s Disease and Healthy Subjects. J. Mol. Neurosci. 2014, 54, 830–836. https://doi.org/10.1007/s12031-014-0428-y
- Miñones-Moyano, E.; Porta, S.; Escaramís, G.; Rabionet, R.; Iraola, S.; Kagerbauer, B.; Espinosa-Parrilla, Y.; Ferrer, I.; Estivill, X.; Martí, E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011, 20, 3067–3078. https://doi.org/10.1093/hmg/ddr210
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of Extracellular miRNA in Cerebrospinal Fluid and Serum from Patients with Alzheimer’s and Parkinson’s Diseases Correlate with Disease Status and Features of Pathology. PLoS ONE 2014, 9, e94839. https://doi.org/10.1371/journal.pone.0094839
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. https://doi.org/10.18632/oncotarget.6158
- Marques, T.M.; Kuiperij, H.B.; Bruinsma, I.B.; van Rumund, A.; Aerts, M.B.; Esselink, R.A.J.; Bloem, B.R.; Verbeek, M.M. MicroRNAs in Cerebrospinal Fluid as Potential Biomarkers for Parkinson’s Disease and Multiple System Atrophy. Mol. Neurobiol. 2017, 54, 7736–7745. https://doi.org/10.1007/s12035-016-0253-0
- Mo, M.; Xiao, Y.; Huang, S.; Cen, L.; Chen, X.; Zhang, L.; Luo, Q.; Li, S.; Yang, X.; Lin, X.; et al. MicroRNA expressing profiles in A53T mutant alpha-synuclein transgenic mice and Parkinsonian. Oncotarget 2017, 8, 15–28. https://doi.org/10.18632/oncotarget.13905
- Oliveira, S.R.; Dionísio, P.A.; Guedes, L.C.; Gonçalves, N.; Coelho, M.; Rosa, M.M.; Amaral, J.D.; Ferreira, J.J.; Rodrigues, C.M.P. Circulating Inflammatory miRNAs Associated with Parkinson’s Disease Pathophysiology. Biomolecules 2020, 10, 945. https://doi.org/10.3390/biom10060945
- Margis, R.; Margis, R.; Rieder, C.R. Identification of blood microRNAs associated to Parkinsońs disease. J. Biotechnol. 2011, 152, 96–101. https://doi.org/10.1016/j.jbiotec.2011.01.023
- Khoo, S.K.; Petillo, D.; Kang, U.J.; Resau, J.H.; Berryhill, B.; Linder, J.; Forsgren, L.; Neuman, L.A.; Tan, A.C. Plasma-Based Circulating MicroRNA Biomarkers for Parkinson’s Disease. J. Park. Dis. 2012, 2, 321–331. https://doi.org/10.3233/JPD-012144
- Botta-Orfila, T.; Morató, X.; Compta, Y.; Lozano, J.J.; Falgàs, N.; Valldeoriola, F.; Pont-Sunyer, C.; Vilas, D.; Mengual, L.; Fernández, M.; et al. Identification of blood serum micro-RNAs associated with idiopathic andLRRK2Parkinson’s disease. J. Neurosci. Res. 2014, 92, 1071–1077. https://doi.org/10.1002/jnr.23377
- Vallelunga, A.; Ragusa, M.; Di Mauro, S.; Iannitti, T.; Pilleri, M.; Biundo, R.; Weis, L.; Di Pietro, C.S.; De Iuliis, A.; Nicoletti, A.; et al. Identification of circulating microRNAs for the differential diagnosis of Parkinson’s disease and Multiple System Atrophy. Front. Cell. Neurosci. 2014, 8, 156. https://doi.org/10.3389/fncel.2014.00156
- Serafin, A.; Foco, L.; Zanigni, S.; Blankenburg, H.; Picard, A.; Zanon, A.; Giannini, G.; Pichler, I.; Facheris, M.F.; Cortelli, P.; et al. Overexpression of blood microRNAs 103a, 30b, and 29a in L-dopa-treated patients with PD. Neurology 2015, 84, 645–653. https://doi.org/10.1212/wnl.0000000000001258
- Ding, H.; Huang, Z.; Chen, M.; Wang, C.; Chen, X.; Chen, J.; Zhang, J. Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Park. Relat. Disord. 2016, 22, 68–73. https://doi.org/10.1016/j.parkreldis.2015.11.014
- Cao, X.-Y.; Lu, J.-M.; Zhao, Z.-Q.; Li, M.-C.; Lu, T.; An, X.-S.; Xue, L.-J. MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci. Lett. 2017, 644, 94–99. https://doi.org/10.1016/j.neulet.2017.02.045
- Caggiu, E.; Paulus, K.; Mameli, G.; Arru, G.; Sechi, G.P.; Sechi, L.A. Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. Eneurologicalsci 2018, 13, 1–4. https://doi.org/10.1016/j.ensci.2018.09.002
- Chen, L.; Yang, J.; Lü, J.; Cao, S.; Zhao, Q.; Yu, Z. Identification of aberrant circulating miRNAs in Parkinson’s disease plasma samples. Brain Behav. 2018, 8, e00941. https://doi.org/10.1002/brb3.941
- Leggio, L.; Vivarelli, S.; L’episcopo, F.; Tirolo, C.; Caniglia, S.; Testa, N.; Marchetti, B.; Iraci, N. microRNAs in Parkinson’s Disease: From Pathogenesis to Novel Diagnostic and Therapeutic Approaches. Int. J. Mol. Sci. 2017, 18, 2698. https://doi.org/10.3390/ijms18122698
- Xiong, R.; Wang, Z.; Zhao, Z.; Li, H.; Chen, W.; Zhang, B.; Wang, L.; Wu, L.; Li, W.; Ding, J.; et al. MicroRNA-494 reduces DJ-1 expression and exacerbates neurodegeneration. Neurobiol. Aging 2014, 35, 705–714. https://doi.org/10.1016/j.neurobiolaging.2013.09.027
- Choi, D.C.; Yoo, M.; Kabaria, S.; Junn, E. MicroRNA-7 facilitates the degradation of alpha-synuclein and its aggregates by promoting autophagy. Neurosci. Lett. 2018, 678, 118–123. https://doi.org/10.1016/j.neulet.2018.05.009
- Junn, E.; Lee, K.-W.; Jeong, B.S.; Chan, T.W.; Im, J.-Y.; Mouradian, M.M. Repression of alpha-synuclein expression and toxicity by microRNA-7. Proc. Natl. Acad. Sci. USA 2009, 106, 13052–13057. https://doi.org/10.1073/pnas.0906277106
- Fragkouli, A.; Doxakis, E. miR-7 and miR-153 protect neurons against MPP+-induced cell death via upregulation of mTOR pathway. Front. Cell. Neurosci. 2014, 8, 182. https://doi.org/10.3389/fncel.2014.00182
- Kabaria, S.; Choi, D.C.; Chaudhuri, A.D.; Mouradian, M.M.; Junn, E. Inhibition of miR-34b and miR-34c enhances α-synuclein expression in Parkinson’s disease. FEBS Lett. 2015, 589, 319–325. https://doi.org/10.1016/j.febslet.2014.12.014
- Li, J.-Q.; Tan, L.; Yu, J.-T. The role of the LRRK2 gene in Parkinsonism. Mol. Neurodegener. 2014, 9, 47. https://doi.org/10.1186/1750-1326-9-47
- Gilks, W.P.; Abou-Sleiman, P.M.; Gandhi, S.; Jain, S.; Singleton, A.; Lees, A.J.; Shaw, K.; Bhatia, K.P.; Bonifati, V.; Quinn, N.P.; et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet 2005, 365, 415–416. https://doi.org/10.1016/s0140-6736(05)17830-1
- Gehrke, S.; Imai, Y.; Sokol, N.; Lu, B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 2010, 466, 637–641. https://doi.org/10.1038/nature09191
- Dong, X.; He, X.; Yang, L.; Li, Q.; Xu, Y. Inhibition of miR-421 Preserves Mitochondrial Function and Protects against Parkinson’s Disease Pathogenesis via Pink1/Parkin-Dependent Mitophagy. Dis. Markers 2022, 2022, 5186252. https://doi.org/10.1155/2022/5186252
- Zeng, R.; Luo, D.-X.; Li, H.-P.; Zhang, Q.-S.; Lei, S.-S.; Chen, J.-H. MicroRNA-135b alleviates MPP+-mediated Parkinson’s disease in in vitro model through suppressing FoxO1-induced NLRP3 inflammasome and pyroptosis. J. Clin. Neurosci. 2019, 65, 125–133. https://doi.org/10.1016/j.jocn.2019.04.004
- Geng, L.; Zhang, T.; Liu, W.; Chen, Y. miR-494-3p modulates the progression of in vitro and in vivo Parkinson’s disease models by targeting SIRT3. Neurosci. Lett. 2018, 675, 23–30. https://doi.org/10.1016/j.neulet.2018.03.037
- Wang, Y.; Cai, Y.; Huang, H.; Chen, X.; Chen, X.; Chen, X.; Mai, H.; Li, X.; Zhao, J.; Yang, J.; et al. miR-486-3p Influences the Neurotoxicity of a-Synuclein by Targeting the SIRT2 Gene and the Polymorphisms at Target Sites Contributing to Parkinson’s Disease. Cell. Physiol. Biochem. 2018, 51, 2732–2745. https://doi.org/10.1159/000495963
- Thome, A.D.; Harms, A.S.; Volpicelli-Daley, L.A.; Standaert, D.G. microRNA-155 Regulates Alpha-Synuclein-Induced Inflammatory Responses in Models of Parkinson Disease. J. Neurosci. 2016, 36, 2383–2390. https://doi.org/10.1523/jneurosci.3900-15.2016
- Zhou, Y.; Lu, M.; Du, R.-H.; Qiao, C.; Jiang, C.-Y.; Zhang, K.-Z.; Ding, J.-H.; Hu, G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 2016, 11, 28. https://doi.org/10.1186/s13024-016-0094-3
- He, Q.; Wang, Q.; Yuan, C.; Wang, Y. Downregulation of miR-7116-5p in microglia by MPP+sensitizes TNF-α production to induce dopaminergic neuron damage. Glia 2017, 65, 1251–1263. https://doi.org/10.1002/glia.23153
- Hanan, M.; Simchovitz, A.; Yayon, N.; Vaknine, S.; Cohen-Fultheim, R.; Karmon, M.; Madrer, N.; Rohrlich, T.M.; Maman, M.; Bennett, E.R.; et al. A Parkinson’s disease Circ RNA s Resource reveals a link between circ SLC 8A1 and oxidative stress. EMBO Mol. Med. 2020, 12, e11942. https://doi.org/10.15252/emmm.201911942
- Paldor, I.; Madrer, N.; Treidel, S.V.; Shulman, D.; Greenberg, D.S.; Soreq, H. Cerebrospinal fluid and blood profiles of transfer RNA fragments show age, sex, and Parkinson’s disease-related changes. J. Neurochem. 2023, 164, 671–683. https://doi.org/10.1111/jnc.15723
- Kurz, A.; Kumar, R.; Northoff, B.H.; Wenk, C.; Schirra, J.; Donakonda, S.; Höglinger, G.U.; Schwarz, J.; Rozanski, V.; Hübner, R.; et al. Differential expression of gut miRNAs in idiopathic Parkinson’s disease. Park. Relat. Disord. 2021, 88, 46–50. https://doi.org/10.1016/j.parkreldis.2021.05.022
- Pellegrini, C.; Antonioli, L.; Calderone, V.; Colucci, R.; Fornai, M.; Blandizzi, C. Microbiota-gut-brain axis in health and disease: Is NLRP3 inflammasome at the crossroads of microbiota-gut-brain communications? Prog. Neurobiol. 2020, 191, 101806. https://doi.org/10.1016/j.pneurobio.2020.101806
- Fung, T.C.; Olson, C.A.; Hsiao, E.Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 2017, 20, 145–155. https://doi.org/10.1038/nn.4476
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. https://doi.org/10.1016/j.chom.2015.12.005
- Hewel, C.; Kaiser, J.; Wierczeiko, A.; Linke, J.; Reinhardt, C.; Endres, K.; Gerber, S. Common miRNA Patterns of Alzheimer’s Disease and Parkinson’s Disease and Their Putative Impact on Commensal Gut Microbiota. Front. Neurosci. 2019, 13, 113. https://doi.org/10.3389/fnins.2019.00113
- Peck, B.C.; Mah, A.T.; Pitman, W.A.; Ding, S.; Lund, P.K.; Sethupathy, P. Functional Transcriptomics in Diverse Intestinal Epithelial Cell Types Reveals Robust MicroRNA Sensitivity in Intestinal Stem Cells to Microbial Status. J. Biol. Chem. 2017, 292, 2586–2600. https://doi.org/10.1074/jbc.m116.770099
- Dalmasso, G.; Nguyen, H.T.T.; Yan, Y.; Laroui, H.; Charania, M.A.; Ayyadurai, S.; Sitaraman, S.V.; Merlin, D. Microbiota Modulate Host Gene Expression via MicroRNAs. PLoS ONE 2011, 6, e19293. https://doi.org/10.1371/journal.pone.0019293
- Viennois, E.; Chassaing, B.; Tahsin, A.; Pujada, A.; Wang, L.; Gewirtz, A.T.; Merlin, D. Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation. Theranostics 2019, 9, 4542–4557. https://doi.org/10.7150/thno.35282
- Moloney, G.; Viola, M.; Hoban, A.; Dinan, T.; Cryan, J. Faecal microRNAs: Indicators of imbalance at the host-microbe interface? Benef. Microbes 2018, 9, 175–183. https://doi.org/10.3920/bm2017.0013
- Nguyen, H.T.T.; Dalmasso, G.; Müller, S.; Carrière, J.; Seibold, F.; Darfeuille–Michaud, A. Crohn’s Disease–Associated Adherent Invasive Escherichia coli Modulate Levels of microRNAs in Intestinal Epithelial Cells to Reduce Autophagy. Gastroenterology 2014, 146, 508–519. https://doi.org/10.1053/j.gastro.2013.10.021
- Hou, Q.; Huang, Y.; Wang, Y.; Liao, L.; Zhu, Z.; Zhang, W.; Liu, Y.; Li, P.; Chen, X.; Liu, F. Lactobacillus casei LC01 Regulates Intestinal Epithelial Permeability through miR-144 Targeting of OCLN and ZO1. J. Microbiol. Biotechnol. 2020, 30, 1480–1487. https://doi.org/10.4014/jmb.2002.02059
- Rodríguez-Nogales, A.; Algieri, F.; Garrido-Mesa, J.; Vezza, T.; Utrilla, M.P.; Chueca, N.; Garcia, F.; Olivares, M.; Rodríguez-Cabezas, M.E.; Gálvez, J. Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: Impact on microRNAs expression and microbiota composition. Mol. Nutr. Food Res. 2017, 61, 1700144. https://doi.org/10.1002/mnfr.201700144
- Kreuzer-Redmer, S.; Bekurtz, J.C.; Arends, D.; Bortfeldt, R.; Kutz-Lohroff, B.; Sharbati, S.; Einspanier, R.; Brockmann, G.A. Feeding of Enterococcus faecium NCIMB 10415 Leads to Intestinal miRNA-423-5p-Induced Regulation of Immune-Relevant Genes. Appl. Environ. Microbiol. 2016, 82, 2263–2269. https://doi.org/10.1128/aem.04044-15
- Sabharwal, H.; Cichon, C.; Ölschläger, T.A.; Sonnenborn, U.; Schmidt, M.A. Interleukin-8, CXCL1, and MicroRNA miR-146a Responses to Probiotic Escherichia coli Nissle 1917 and Enteropathogenic E. coli in Human Intestinal Epithelial T84 and Monocytic THP-1 Cells after Apical or Basolateral Infection. Infect. Immun. 2016, 84, 2482–2492. https://doi.org/10.1128/iai.00402-16
- Hoban, A.E.; Stilling, R.M.; Moloney, G.M.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; Clarke, G. Microbial regulation of microRNA expression in the amygdala and prefrontal cortex. Microbiome 2017, 5, 102. https://doi.org/10.1186/s40168-017-0321-3
- Zhao, Y.; Lukiw, W.J. Bacteroidetes Neurotoxins and Inflammatory Neurodegeneration. Mol. Neurobiol. 2018, 55, 9100–9107. https://doi.org/10.1007/s12035-018-1015-y
- Yadav, R.; Ramaswamy, P.; Pal, P.K.; Christopher, R. Clinical application of circulating micrornas in parkinson’s disease: The challenges and opportunities as diagnostic biomarker. Ann. Indian Acad. Neurol. 2020, 23, 84–97. https://doi.org/10.4103/aian.aian_440_19
- Goh, S.Y.; Chao, Y.X.; Dheen, S.T.; Tan, E.-K.; Tay, S.S.-W. Role of MicroRNAs in Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 5649. https://doi.org/10.3390/ijms20225649
