Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Exosomes have been proposed as prospective “Trojan horse” nanocarriers of anticancer theranostics owing to their biocompatibility, increased stability, permeability, negligible immunogenicity, prolonged circulation time, and high loading capacity.
- exosomes
- brain cancer
- glioblastoma
1. Physicochemical Characteristics of Exosomes
Nowadays, scientists provide a big arsenal of various natural nanosystems that utilize a spectrum of natural materials, including proteins, lipids, polysaccharides, synthetic polymers, and inorganic materials, differing in shapes, sizes, structures, and compositions [1][2][3][4][5]. Owing to the combination of advantageous physicochemical and biological characteristics of natural components, the aforementioned NPs have been recently assessed as auspicious fundamental materials for cancer therapy. In particular, natural-derived nanostructures possess desired properties, such as enhanced biocompatibility, biodegradability, non-immunogenicity, and superior safety profiles in contrast to the majority of synthetic NPs. Moreover, they own functional groups that promote their reaction with chemicals enabling their modification, thus leading to the achievement of more effective final formulations [2][4][6].
Additionally, due to the fundamental role of toxicology evaluations in the further clinical outcome assessment of particulate nano drugs [7], the propitious cellular response to natural NPs could their rapid exploitation in clinical tests, and subsequently, in various medicinal applications instead of synthetic alternative solutions. An extensive list of anticancer nanoparticle formulations containing resveratrol and curcumin (Cur) recording their physicochemical characteristics, the type of cancer, and crucial effects is reviewed in detail [8].
Despite the progress in this field, several concerns have been raised regarding the use of these NPs, relating to their bioavailability, expense, undesirable outcomes, including cytotoxic effects on diverse cells [9][10], biocompatibility, and a very limited half-life in vivo, which is revealed by the swift removal from the systemic circulation [11].
Exosomes are natural NPs able to transport and distribute a spectrum of proteins as well as some nucleic acids, mainly RNA, whereas they are also liable to subjection to the engineering process in order to ameliorate their effectiveness against tumor progression [12]. Interestingly, these nano-scale vesicles originated from membrane invaginations and originally participated in the communication between cells; they display penetration capacity through the BBB, which is an advantage that enables their utilization as vehicles for the transportation of administered therapeutic agents for brain diseases [13].
Exosome applications have emerged as an auspicious nanotechnology field expanded to several diseases, therefore contributing to both diagnosis and therapy. Owing to their innate properties, exosomes perform efficient drug transportation, exhibit high biocompatibility inside the organism, permeate easily through physiological barriers, and cause negligible or mild side effects, thus diminishing the distance until clinical tests and translational medicine [13]. A recent review highlights the attempts documented at drug transportation utilizing exosomes destined for neurodegenerative diseases and brain cancer therapy, while it also synopsizes the possible hindrances and future perspectives that will emerge in clinical translation [13].
Notably, exosomes regarding their properties excel beyond other NPs because of their remarkable biocompatibility, negligible immunogenicity, sufficient stability, protracted circulation half-life, recognition of their targets, and ability to cross physical barriers such as the BBB [14]. Additionally, their capability to carry active biomolecules with therapeutic and diagnostic potential as well as their intrinsic propensity for bioengineering has been greatly recognized nowadays.
2. Exosome Isolation
Taking the physical and chemical properties of extracellular vehicles (EVs) into consideration, exosome isolation from body fluids was attempted using several methods that aimed to ameliorate the yield, purity, structural integrity, and functionality of exosomes, leading indeed to a final preparation with altered parameters [15]. Based on the applied isolation method, the exosomal purity and the physicochemical characteristics could be affected [13]. The methods that have evolved to enhance the aforementioned exosomal parameters include several size-based techniques, such as tangential flow filtration (TFF) [16], ultrafiltration apparatus [17], size-exclusion chromatography (SEC) [18], polymer precipitation [19][20], ultracentrifugation [21], and microfluidics techniques [22][23].
An acoustic nano-filter could utilize ultrasound standing waves to apply electromagnetic wave forces on the biological liquid and permit the vesicles’ segregation depending on their mechanical properties such as their dimensions and density [24]. Currently, another standard protocol was reported involving a number of purification steps to achieve a suitable exosomal preparation [25], although the final batch-to-batch purity, as well as the sample concentration, is not sufficiently reproducible.
Concerning the employment of exosomes in medical applications and clinical use, a protocol that ensures the production of a sufficient amount of particles is required. Due to the high productivity of exosomal mass, which is necessary for clinical utility, TFF is regarded as the most convenient method, since it provides a higher yield with minor non-exosomal component contamination that usually occurs in serial centrifugations. In addition, high exosomal reproducibility has urged some corporations, involved in exosome commerce, to install TFF-based exosome manufacturing facilities [26].
According to a worldwide International Society for Extracellular Vesicles (ISEV) survey [27], ultracentrifugation (UC)-based exosome isolation is considered to be the most ordinarily utilized method [21][28][29][30][31][32][33]. Exosome isolation in gliomas has been described in detail [34]. By highlighting the lack of an established method to receive a homogeneous exosomal preparation, an exosome comparative quantification approach in terms of particles and protein concentration (μg/mL) was performed, depending on the isolation method, cellular origin, and source availability [34]. The benefits and drawbacks of exosomal purification methods have been recently reviewed [35][36].
3. Exosome Size
EVs differ in size depending on their type, ranging from 30 to 5000 nm, including exosomes, exomeres, microvesicles (MVs) or ectosomes, and apoptotic bodies, as well as in the mechanism of their biogenesis [37]. Even though the categorization of EVs, relying on exosomal size, is ceaselessly progressing [38], they are normally classified into four main subcategories containing
Other studies have reported that EVs are classified into three major subclasses including MVs or ectosomes, apoptotic bodies, and exosomes [41][42]. Their smaller size and unified shape permit exosomes to successfully elude the elimination by the mononuclear phagocyte system, leading to a prolonged circulation duration but also suggesting their supremacy concerning the communication between cells [37].
4. Exosome Stability
Regarding their stability, exosomes are proved to be adequately stable inside the majority of the body fluids, thus protecting the encapsulated biomolecules from catabolic processes. When the cells are under pathological conditions, they are subjected to alterations mirrored in the secretion of biological exosomal constituents. In the case of a differentiated exosomal cargo, this could be detected and correlated to the transcriptome, proteome, and lipidome analyses to gain insights. Consequently, the determination of fluctuations in the content of certain selected components will contribute to the exploitation of exosomes in the biomarker field [43].
Although exosomes represent a favorable cell-free therapy, a drawback regarding their storage stability exists, since they cannot be stored for an extended time period. Henceforth, to confront this problem, exosomes’ maintenance technology with the goal of keeping intact their biological potential and rendering them suitable for shuttle and clinical performance should be taken into consideration. Currently, the utilized shielding methods mostly comprise freezing, freeze-drying, and spray-drying [44]. Upon their preparation, when exosomes are lyophilized, they become more stable at higher temperatures [40].
Due to the distinct complexity of exosomes, their dimension range, and natural incompatibilities during their assembly, the inherent hazards that follow the biogenesis process are more than those of the synthetic production [45][46]. These properties reflect on the variability of their reproducible load with therapeutic drugs, which is something that requires suitable technological approaches [45].
5. Exosome Biological Properties
Exosomes own unique and inspiring properties that have motivated scientists to employ them for therapeutic approaches against many diseases, including degenerative disorders, cancers [37][38], and, mainly, brain cancer. Due to these suitable properties, their therapeutic benefits, and unelucidated functions, exosomes elicit the inquisitiveness of researchers to develop them as drug transportation systems [47].
Their advantageous properties are enumerated as the adequate stability in both physiological and pathological conditions, their small and relatively homogeneous dimensions, their ability for intracellular distribution of selected load through the fusion process of membranes, the capability to traverse physiological barriers, such as the BBB, the absence of immunogenicity and the autologous use permitting personalized medicine [5]. Another benefit that exosomes possess is attributed to their bio-originated membrane, which is inactive in the generation of the named “protein corona”, which is the abundant proteinaceous coating that usually cover NPs when they interact with proteins that are adsorbed to their surfaces in various biological processes [48][49].
On the contrary, among the disadvantages are the requirement of standardized protocols for their isolation and purification, the necessity of sufficient characterization of cell provenance, the undesirable outcomes attributed to the exosome constituents themselves, and the absence of standardized mass production protocols for clinical applications [5].
Because of the heterogeneity of small EVs and the existence of non-vesicular extracellular material, a debate has emerged concerning the composition and functional properties of exosomes. By utilizing high-resolution density gradient fractionation and direct immunoaffinity capture techniques, the constituents harbored by exosomes and other non-vesicle material, including nucleic acids and proteins, were precisely characterized [50].
Exosomes participate in a spectrum of functions because of the membrane-connected proteins, e.g., tetraspanins (CD63, CD81, CD9, CD37, CD53, and CD82), intercellular adhesion molecule-1 (ICAM-1), integrins, intracellular transportation and membrane fusion proteins, such as Ras-associated binding (Rab) and Annexins, proteins related to multi-vesicular bodies (MVBs) and the endocytic pathway, i.e., the endosomal sorting complex needed for transport (ESCRT). For instance, Alix, tumor susceptibility gene 101 (TSG101), proteins mainly participate in vesicle trafficking, such as cell surface receptors and antigen presentation molecules, such as major histocompatibility complex I and II (MHC-I, MHC-II), human leukocyte antigen-G (HLA-G) [51] and, in addition, other proteins implicated in long-distance signal transduction, such as cytokines [52], hormones [53], growth and transcription factors [54], and heat shock proteins (HSPs) [55][56]. Moreover, the protein content of the membrane depends on the cell type and the methodology of purification, as portrayed in the record of databases ExoCarta [28][57], Vesiclepedia [58] and EVpedia [59]. An abundant depository completes the exosomal composition, including cholesterol, sphingomyelin, phosphatidylcholine, phosphatidylserine, phosphatidylethanolamine, phosphatidylinositol, ganglioside GM3, prostaglandins, and lyso-bisphosphatidic acid [60][61].
There is evidence that exosomes comprise or express a variety of bioactive molecules including proteins, RNAs (mRNA, microRNA, and other non-coding RNAs), DNAs (mitochondrial DNA (mtDNA), double-stranded DNA (dsDNA), single-stranded DNA and viral DNA, lipids, amino acids, and metabolites. These miscellaneous constituents exert important roles, such as in intercellular signaling and regulating neighboring or remote cellular microenvironments [62][63][64]. Moreover, to obtain insights into exosomes composition, small EV exosomes, and non-vesicular components, a combination of the direct immune affinity capture method targeting the classical exosome marker tetraspanin with high-resolution density gradient fractionation was applied [35]. Although exosomes and non-vesicular components were found to bear different proteins and RNAs, they did not possess microRNA (miRNA) processing proteins, such as Argonautes (Agos), glycolytic enzymes, or cytoskeletal proteins. Moreover, others reported that exosomes did not contain double-stranded DNA (dsDNA) or DNA-binding histones, which are typically found in exosomes, suggesting that dsDNA or DNA-binding histones are released through an exosome-unmediated mechanism. These outcomes highlight the necessity for a revaluation of the exosome composition by an in-depth analysis to elucidate exosomal heterogeneity [35]. One idiosystatic and noteworthy property is that endogenous exosomes operate as “signalosomes” inasmuch as they transmit signals through ligands or adhesion molecules localized on the exosomal membrane or enclosed in the interior of the exosome. This communication carried out by the intervention of the exosome has several unique features [65]. Specifically, exosomes exchange various substances between the cells without contact and, as a result, participate in various ways in cell communication and signal transduction processes, thus comprising autocrine, endocrine, juxtracrine, paracrine, or in distance transmission [66].
The existence of exosomes in biofluids may exhibit certain diagnostic and therapeutic properties. For instance, the extensive investigation of circular RNAs (circRNAs) present in exosomes circulating in biological fluids, such as blood, or cerebrospinal fluid (CSF) may contribute to discovering the fundamental mechanisms of the progress of neuropsychiatric diseases [43]. Furthermore, miRNAs transported by serum exosomes are useful for the prognosis and diagnosis of spinal cord injury (SCI) [67]. The recognition of the therapeutic properties of exosomes renders them as candidates for utilization in clinical applications that are dependent on the three following features: the kind of vesicle and load and/or the type of surface modifications or functional encapsulated molecules [12].
6. Exosome Biogenesis and Internalization
According to the exosome biogenesis pathway, exosomes are derived from the endosomal membrane, creating a toward-the-inside budding, which leads to the generation of MVBs. Subsequently, MVBs possess two fates, either degradation into the lysosomes or fusion with the plasma membrane, and the subsequent externalization of exosomes.
Exosomes are supposed to constitute a homogeneous ensemble of vesicles that originate from the inner budding of the MVBs membrane. After MVBs fusion with the cellular membrane, the secretion of exosomes occurs, displaying the same membrane orientation as the parent cells [16]. For comparison, MVs constitute an almost heterogeneous group of vesicles shaped by exterior folding followed by disruption of the cell membrane, which could be controlled by some membrane lipid micro areas and some regulatory proteins, such as ADP Ribosylation Factor 6 (ARF6). Thus, both exosomes and MVs can be assumed as “signalosomes” implicated in diverse biological processes (Figure 1).
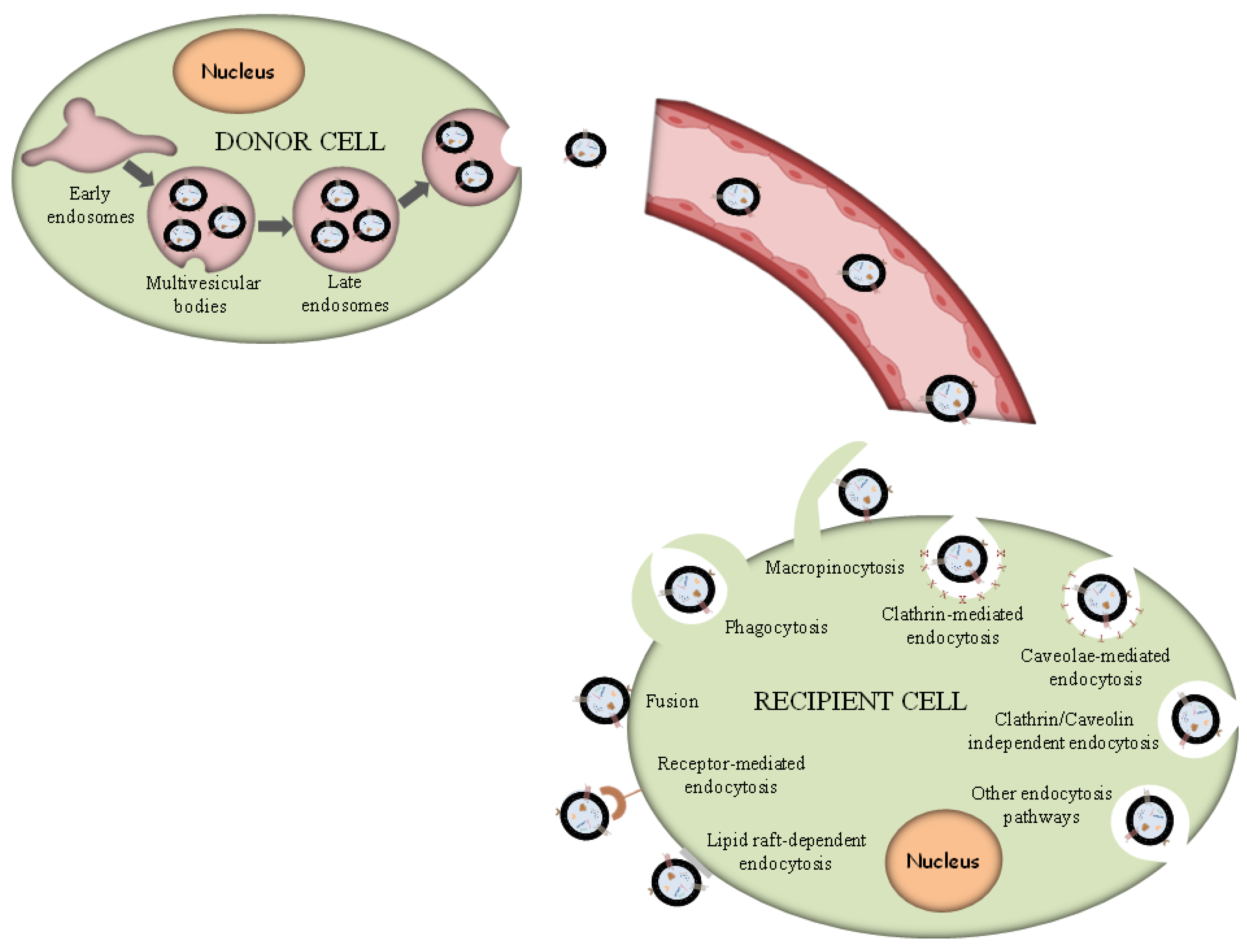
Figure 1. Exosome biogenesis and different uptake pathways.
After their exocytosis, exosomes migrate into the extracellular matrix (ECM), where exosomal surface proteins assist in the detection of the target cells for the subsequent uptake [68]. Exosomes interact with particular receptors of the recipient cells by the intervention of integrins, tetraspanins and intercellular adhesion molecules, which subsequently internalize exosomes through the same conventional means following the uptake of various molecules into the cells, including:
- (i)
-
Phagocytosis. Phagocytosis is almost a similar internalization process to macropinocytosis where exosomes, along with some extracellular fluids, are phagocytosed. This process is common and adopted by both phagocytic cells, i.e., macrophages and dendritic cells (DCs) and non-phagocytic cells, i.e., T cells [69].
- (ii)
-
Macropinocytosis. Macropinocytosis is a process in which the deformation of the plasma membrane leads to the formation of protrusions that surround the extracellular fluid and exosomes, therefrom uptaking exosomes. This complicated process is mediated by many factors, since it relies on Ras-related C3 botulinum toxun substrate 1 (Rac1), actin and cholesterol and it requires Na+/H+ exchange [70].
- (iii)
-
Clathrin/caveolin-mediated endocytosis. The Clathrin protein shapes a basket knitted-like structure around the exosomes for their internalization. The plasma membrane of the target cells creates an internal folding, which is followed by squeezing off the clathrin-coated vesicle from the membrane. Subsequently, the exosome releases all its cargo in the targets cell’s endosomes to implement particular functions [71].
- (iv)
-
Internalization through lipid raft. The endocytosis process may be also mediated by caveolin-1, similar to the clathrin-dependent way, whose clusters in plasma membrane are configured in rafts. The invagination of the plasma membrane named caveolae is abundant in caveolin 1, glycolipids, and cholesterol [72], and
- (v)
-
Direct fusion with the plasma membrane. In this fusion process, the Lysosomal-Associated Membrane Protein 1 (LAMP-1) and glycoprotein type I transmembrane protein are implicated primarily residing across lysosomal membranes, and in certain cases, it can be expressed across the plasma membrane of the cell, and the integrins or tetraspanins [73][74].
The internalization of exosomes through the receptor-mediated endocytosis process has been reviewed extensively [70], and a number of receptor–ligand complexes listed are considered to facilitate exosomal internalization, while several other proteins have been identified as contributors in this endocytosis process of exosomes through protein–protein interactions implicated in the exosomal uptake. Apart from the detailed presented receptor/ligand complexes, other receptor-mediated endocytosis ligands and receptors have been also identified, but still, further evaluation is required to elucidate their role in exosomal internalization (Figure 1).
7. Cancer Cell-Derived Exosomes
Cancer cell-derived exosomes are pivotally involved in tumor-linked processes including proliferation, metastasis, and immune regulation [37][38], whereas they might be employed to surveil the progress of disease [39], serving as selected diagnostic markers. Although all cells possess the ability to generate exosomes, tumor-derived exosomes (TdEX) seem to display distinguishing characteristics from exosomes secreted by normal cells, being mightily immunosuppressive [75][76]. TdEX can transmit signals between cells or transport various biological active matter toward the recipient cells, in this way triggering alterations of their cellular features [75][76][77]. Another important property that should be taken into consideration is that exosomes and, especially, those originating from tumor cells are considered to preferentially perform delivery to tumors, which is a phenomenon attributed to their homotypic features. Due to these properties, the TdEX has received growing interest to be considered as a potential clinical biomarker, and they may constitute particular vehicles for specific cancer targeting. Moreover, the pharmacokinetic properties of naive exosomes and factors contributing their intracellular “luck or destiny” have been recently summarized [65].
Cancer cells-secreted exosomes convey pathophysiological features and function as intermediates of tumor progression and distant lesions genesis [48][49][50]. Several in vitro studies propose that in the micro-surrounding of a tumor, cancer cell-derived exosomes probably attack other neoplastic cells in preference to physiological cells, therefore employing them as selective distribution vehicles [78]. In addition, cancer cell-derived exosomes carry various DNA, including genomic, cytoplasmic, and mitochondrial DNA as particular cancer cargo, alongside with other cellular molecules such as RNA, proteins, and lipid rafts. All the previously mentioned biomolecules, that are pleiotropically functional in the receiver cell, contribute to the alteration of the tumor microenvironment, angiogenesis, metastasis, and immunity avoidance [79].
Exosomes are responsible for their decisive role in cancer spreading because of their mediation in DNA horizontal transfer, comprising specific differentiations attributed to tumors. These differentiations occur by transmitting DNA from cancer cells to healthy ones, henceforth reflecting the transformation of the receiver cell to a condition premalignant or malignant. Exosome-derived miRNAs may also be considered as candidate biomarkers for glioma, although additional scientific attempts are required to promote the investigation of clinically helpful biomarkers for gliomas [80].
In brain cancer, a resistant cancer type in immune cell recruitment, exosomes originated from DCs have been considered as a favorable treatment strategy against glioma, applied in a mice model, and this immune-related and exosome-based approach constituted an unconventional therapy for GBM [81]. Another noteworthy property of exosomes is that they can traverse through an uninjured BBB and anatomical compartments through transcytosis, which is the common strategy of multicellular organisms utilized to selectively transport vesicular macromolecules between two surroundings without altering their unique composition [82].
In contrast to the natural ones, exosomes derived from desired design and modifications could eliminate the restrictions derived from natural exosomes, such as lack of target recognition capability, insufficient half-life in circulation, and a small quantity of functional molecules. Similarly, exosomes profit concomitantly from their content in surface-decorated and encapsulated molecules [83].
Many attempts have been made to clarify the properties of TdEXs as sources of tumor antigens and immune adjuvants. Nevertheless, several statements have proposed TdEXs to be involved with pro-tumorigenic roles. The complicated functionalities of TdEXs must be investigated exhaustively, and an in-depth exploration into TdEX biogeneration and identification will expand their usefulness by permitting the TdEXs optimization reflecting on the alleviation of their adverse features and amplifying their potency [35].
This entry is adapted from the peer-reviewed paper 10.3390/pharmaceutics15051439
References
- Davatgaran-Taghipour, Y.; Masoomzadeh, S.; Farzaei, M.H.; Bahramsoltani, R.; Karimi-Soureh, Z.; Rahimi, R.; Abdollahi, M. Polyphenol Nanoformulations for Cancer Therapy: Experimental Evidence and Clinical Perspective. Int. J. Nanomed. 2017, 12, 2689–2702.
- Choi, D.G.; Venkatesan, J.; Shim, M.S. Selective Anticancer Therapy Using Pro-Oxidant Drug-Loaded Chitosan–Fucoidan Nanoparticles. Int. J. Mol. Sci. 2019, 20, 3220.
- Abdelaziz, H.M.; Gaber, M.; Abd-Elwakil, M.M.; Mabrouk, M.T.; Elgohary, M.M.; Kamel, N.M.; Kabary, D.M.; Freag, M.S.; Samaha, M.W.; Mortada, S.M.; et al. Inhalable Particulate Drug Delivery Systems for Lung Cancer Therapy: Nanoparticles, Microparticles, Nanocomposites and Nanoaggregates. J. Control. Release 2018, 269, 374–392.
- Curcio, M.; Diaz-Gomez, L.; Cirillo, G.; Nicoletta, F.P.; Leggio, A.; Iemma, F. Dual-Targeted Hyaluronic Acid/Albumin Micelle-Like Nanoparticles for the Vectorization of Doxorubicin. Pharmaceutics 2021, 13, 304.
- de la Torre, P.; Pérez-Lorenzo, M.J.; Alcázar-Garrido, Á.; Flores, A.I. Cell-Based Nanoparticles Delivery Systems for Targeted Cancer Therapy: Lessons from Anti-Angiogenesis Treatments. Molecules 2020, 25, 715.
- AlMalki, F.A.; Hassan, A.M.; Klaab, Z.M.; Abdulla, S.; Pizzi, A. Tannin Nanoparticles (NP99) Enhances the Anticancer Effect of Tamoxifen on ER+ Breast Cancer Cells. J. Renew. Mater. 2021, 9, 2077–2092.
- Hua, S.; de Matos, M.B.C.; Metselaar, J.M.; Storm, G. Current Trends and Challenges in the Clinical Translation of Nanoparticulate Nanomedicines: Pathways for Translational Development and Commercialization. Front. Pharmacol. 2018, 9, 790.
- Ion, D.; Niculescu, A.G.; Păduraru, D.N.; Andronic, O.; Mușat, F.; Grumezescu, A.M.; Bolocan, A. An Up-to-Date Review of Natural Nanoparticles for Cancer Management. Pharmaceutics 2022, 14, 18.
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383.
- Hood, J.L. Post Isolation Modification of Exosomes for Nanomedicine Applications. Nanomedicine 2016, 11, 1745–1756.
- Hood, J.L.; Wickline, S.A. A Systematic Approach to Exosome-Based Translational Nanomedicine. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2012, 4, 458–467.
- You, B.; Xu, W.; Zhang, B. Engineering Exosomes: A New Direction for Anticancer Treatment. Am. J. Cancer Res. 2018, 8, 1332–1342.
- Rehman, F.U.; Liu, Y.; Zheng, M.; Shi, B. Exosomes Based Strategies for Brain Drug Delivery. Biomaterials 2023, 293, 121949.
- Samanta, S.; Rajasingh, S.; Drosos, N.; Zhou, Z.; Dawn, B.; Rajasingh, J. Exosomes: New Molecular Targets of Diseases. Acta Pharmacol. Sin. 2018, 39, 501–513.
- Jia, S.; Zocco, D.; Samuels, M.L.; Chou, M.F.; Chammas, R.; Skog, J.; Zarovni, N.; Momen-Heravi, F.; Kuo, W.P. Emerging Technologies in Extracellular Vesicle-Based Molecular Diagnostics. Expert Rev. Mol. Diagn. 2014, 14, 307–321.
- Yuana, Y.; Levels, J.; Grootemaat, A.; Sturk, A.; Nieuwland, R. Co-Isolation of Extracellular Vesicles and High-Density Lipoproteins Using Density Gradient Ultracentrifugation. J. Extracell. Vesicles 2014, 3, 23262.
- Cheruvanky, A.; Zhou, H.; Pisitkun, T.; Kopp, J.B.; Knepper, M.A.; Yuen, P.S.T.; Star, R.A. Rapid Isolation of Urinary Exosomal Biomarkers Using a Nanomembrane Ultrafiltration Concentrator. Am. J. Physiol. Physiol. 2007, 292, F1657–F1661.
- Böing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-Step Isolation of Extracellular Vesicles by Size-Exclusion Chromatography. J. Extracell. Vesicles 2014, 3, 23430.
- Deregibus, M.C.; Figliolini, F.; D’antico, S.; Manzini, P.M.; Pasquino, C.; De Lena, M.; Tetta, C.; Brizzi, M.F.; Camussi, G. Charge-Based Precipitation of Extracellular Vesicles. Int. J. Mol. Med. 2016, 38, 1359–1366.
- Niu, Z.; Pang, R.T.K.; Liu, W.; Li, Q.; Cheng, R.; Yeung, W.S.B. Polymer-Based Precipitation Preserves Biological Activities of Extracellular Vesicles from an Endometrial Cell Line. PLoS ONE 2017, 12, e0186534.
- Lobb, R.J.; Becker, M.; Wen Wen, S.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized Exosome Isolation Protocol for Cell Culture Supernatant and Human Plasma. J. Extracell. Vesicles 2015, 4, 27031.
- Chen, C.; Skog, J.; Hsu, C.-H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab Chip 2010, 10, 505–511.
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804.
- Lee, K.; Shao, H.; Weissleder, R.; Lee, H. Acoustic Purification of Extracellular Microvesicles. ACS Nano 2015, 9, 2321–2327.
- Gu, Y.; Li, M.; Wang, T.; Liang, Y.; Zhong, Z.; Wang, X.; Zhou, Q.; Chen, L.; Lang, Q.; He, Z.; et al. Lactation-Related MicroRNA Expression Profiles of Porcine Breast Milk Exosomes. PLoS ONE 2012, 7, e43691.
- Busatto, S.; Vilanilam, G.; Ticer, T.; Lin, W.-L.; Dickson, D.; Shapiro, S.; Bergese, P.; Wolfram, J. Tangential Flow Filtration for Highly Efficient Concentration of Extracellular Vesicles from Large Volumes of Fluid. Cells 2018, 7, 273.
- Gardiner, C.; Vizio, D.D.; Sahoo, S.; Théry, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques Used for the Isolation and Characterization of Extracellular Vesicles: Results of a Worldwide Survey. J. Extracell. Vesicles 2016, 5, 32945.
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human Colon Cancer Cell Line LIM1863-Derived Exosomes. Methods 2012, 56, 293–304.
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Sano, K.; Kato, K.; Yamashita, T.; Imai, T.; Saji, H.; Takakura, Y. Quantitative Analysis of Tissue Distribution of the B16BL6-Derived Exosomes Using a Streptavidin-Lactadherin Fusion Protein and Iodine-125-Labeled Biotin Derivative After Intravenous Injection in Mice. J. Pharm. Sci. 2015, 104, 705–713.
- Smyth, T.; Kullberg, M.; Malik, N.; Smith-Jones, P.; Graner, M.W.; Anchordoquy, T.J. Biodistribution and Delivery Efficiency of Unmodified Tumor-Derived Exosomes. J. Control. Release 2015, 199, 145–155.
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as Drug Delivery Vehicles for Parkinson’s Disease Therapy. J. Control. Release 2015, 207, 18–30.
- Didiot, M.-C.; Hall, L.M.; Coles, A.H.; Haraszti, R.A.; Godinho, B.M.; Chase, K.; Sapp, E.; Ly, S.; Alterman, J.F.; Hassler, M.R.; et al. Exosome-Mediated Delivery of Hydrophobically Modified SiRNA for Huntingtin MRNA Silencing. Mol. Ther. 2016, 24, 1836–1847.
- Faruqu, F.N.; Wang, J.T.-W.; Xu, L.; McNickle, L.; Chong, E.M.-Y.; Walters, A.; Gurney, M.; Clayton, A.; Smyth, L.A.; Hider, R.; et al. Membrane Radiolabelling of Exosomes for Comparative Biodistribution Analysis in Immunocompetent and Immunodeficient Mice—A Novel and Universal Approach. Theranostics 2019, 9, 1666–1682.
- Romano, E.; Netti, P.A.; Torino, E. Exosomes in Gliomas: Biogenesis, Isolation, and Preliminary Applications in Nanomedicine. Pharmaceuticals 2020, 13, 319.
- Nam, G.H.; Choi, Y.; Kim, G.B.; Kim, S.; Kim, S.A.; Kim, I.S. Emerging Prospects of Exosomes for Cancer Treatment: From Conventional Therapy to Immunotherapy. Adv. Mater. 2020, 32, 2002440.
- Zhang, Y.; Li, J.; Gao, W.; Xie, N. Exosomes as Anticancer Drug Delivery Vehicles: Prospects and Challenges. Front. Biosci. Landmark 2022, 27, 293.
- Rezaie, J.; Feghhi, M.; Etemadi, T. A Review on Exosomes Application in Clinical Trials: Perspective, Questions, and Challenges. Cell Commun. Signal. 2022, 20, 145.
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750.
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding Microvesicles: Artefacts No More. Trends Cell Biol. 2009, 19, 43–51.
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606.
- Patil, A.A.; Rhee, W.J. Exosomes: Biogenesis, Composition, Functions, and Their Role in Pre-Metastatic Niche Formation. Biotechnol. Bioprocess Eng. 2019, 24, 689–701.
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2021, 8, 2003505.
- Zhuo, C.-J.; Hou, W.-H.; Jiang, D.-G.; Tian, H.-J.; Wang, L.-N.; Jia, F.; Zhou, C.-H.; Zhu, J.-J. Circular RNAs in Early Brain Development and Their Influence and Clinical Significance in Neuropsychiatric Disorders. Neural Regen. Res. 2020, 15, 817.
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934.
- Armstrong, J.P.K.; Stevens, M.M. Strategic Design of Extracellular Vesicle Drug Delivery Systems. Adv. Drug Deliv. Rev. 2018, 130, 12–16.
- Kibria, G.; Ramos, E.K.; Wan, Y.; Gius, D.R.; Liu, H. Exosomes as a Drug Delivery System in Cancer Therapy: Potential and Challenges. Mol. Pharm. 2018, 15, 3625–3633.
- Peng, H.; Ji, W.; Zhao, R.; Yang, J.; Lu, Z.; Li, Y.; Zhang, X. Exosome: A Significant Nano-Scale Drug Delivery Carrier. J. Mater. Chem. B 2020, 8, 7591–7608.
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977.
- Vahabi, A.; Rezaie, J.; Hassanpour, M.; Panahi, Y.; Nemati, M.; Rasmi, Y.; Nemati, M. Tumor Cells-Derived Exosomal CircRNAs: Novel Cancer Drivers, Molecular Mechanisms, and Clinical Opportunities. Biochem. Pharmacol. 2022, 200, 115038.
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Baldelli Bombelli, F.; Dawson, K.A. Physical–Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534.
- Lunova, M.; Smolková, B.; Lynnyk, A.; Uzhytchak, M.; Jirsa, M.; Kubinová, Š.; Dejneka, A.; Lunov, O. Targeting the MTOR Signaling Pathway Utilizing Nanoparticles: A Critical Overview. Cancers 2019, 11, 82.
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18.
- Lo Cicero, A.; Stahl, P.D.; Raposo, G. Extracellular Vesicles Shuffling Intercellular Messages: For Good or for Bad. Curr. Opin. Cell Biol. 2015, 35, 69–77.
- Qu, Y.; Franchi, L.; Nunez, G.; Dubyak, G.R. Nonclassical IL-1β Secretion Stimulated by P2X7 Receptors Is Dependent on Inflammasome Activation and Correlated with Exosome Release in Murine Macrophages. J. Immunol. 2007, 179, 1913–1925.
- Phoonsawat, W.; Aoki-Yoshida, A.; Tsuruta, T.; Sonoyama, K. Adiponectin Is Partially Associated with Exosomes in Mouse Serum. Biochem. Biophys. Res. Commun. 2014, 448, 261–266.
- Chen, L.; Chen, R.; Kemper, S.; Charrier, A.; Brigstock, D.R. Suppression of Fibrogenic Signaling in Hepatic Stellate Cells by Twist1-Dependent MicroRNA-214 Expression: Role of Exosomes in Horizontal Transfer of Twist1. Am. J. Physiol. Liver Physiol. 2015, 309, G491–G499.
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of Exosomal Proteins, RNA and Lipids. Nucleic Acids Res. 2012, 40, D1241–D1244.
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a Resource for Exosomal Research. J. Extracell. Vesicles 2012, 1, 18374.
- Kim, D.-K.; Kang, B.; Kim, O.Y.; Choi, D.; Lee, J.; Kim, S.R.; Go, G.; Yoon, Y.J.; Kim, J.H.; Jang, S.C.; et al. EVpedia: An Integrated Database of High-Throughput Data for Systemic Analyses of Extracellular Vesicles. J. Extracell. Vesicles 2013, 2, 20384.
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome Lipidomics Unravels Lipid Sorting at the Level of Multivesicular Bodies. Biochimie 2007, 89, 205–212.
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; De Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes Account for Vesicle-Mediated Transcellular Transport of Activatable Phospholipases and Prostaglandins. J. Lipid Res. 2010, 51, 2105–2120.
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579.
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of MRNAs and MicroRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659.
- Skog, J.; Würdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476.
- Shao, J.; Zaro, J.; Shen, Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int. J. Nanomed. 2020, 15, 9355–9371.
- EL Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J.A. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357.
- Shaimardanova, A.; Solovyeva, V.; Chulpanova, D.; James, V.; Kitaeva, K.; Rizvanov, A. Extracellular Vesicles in the Diagnosis and Treatment of Central Nervous System Diseases. Neural Regen. Res. 2020, 15, 586.
- Heusermann, W.; Hean, J.; Trojer, D.; Steib, E.; von Bueren, S.; Graff-Meyer, A.; Genoud, C.; Martin, K.; Pizzato, N.; Voshol, J.; et al. Exosomes Surf on Filopodia to Enter Cells at Endocytic Hot Spots, Traffic within Endosomes, and Are Targeted to the ER. J. Cell Biol. 2016, 213, 173–184.
- Wu, Y.; Wu, W.; Wong, W.M.; Ward, E.; Thrasher, A.J.; Goldblatt, D.; Osman, M.; Digard, P.; Canaday, D.H.; Gustafsson, K. Human Γδ T Cells: A Lymphoid Lineage Cell Capable of Professional Phagocytosis. J. Immunol. 2009, 183, 5622–5629.
- Gonda, A.; Kabagwira, J.; Senthil, G.N.; Wall, N.R. Internalization of Exosomes through Receptor-Mediated Endocytosis. Mol. Cancer Res. 2019, 17, 337–347.
- Tian, T.; Zhu, Y.-L.; Zhou, Y.-Y.; Liang, G.-F.; Wang, Y.-Y.; Hu, F.-H.; Xiao, Z.-D. Exosome Uptake through Clathrin-Mediated Endocytosis and Macropinocytosis and Mediating MiR-21 Delivery. J. Biol. Chem. 2014, 289, 22258–22267.
- Nabi, I.R.; Le, P.U. Caveolae/Raft-Dependent Endocytosis. J. Cell Biol. 2003, 161, 673–677.
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455.
- Andreu, Z.; Yáñez-MÃ3, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5, 442.
- D’Asti, E.; Garnier, D.; Lee, T.H.; Montermini, L.; Meehan, B.; Rak, J. Oncogenic Extracellular Vesicles in Brain Tumor Progression. Front. Physiol. 2012, 3, 294.
- Whiteside, T.L. Immune Modulation of T-Cell and NK (Natural Killer) Cell Activities by TEXs (Tumour-Derived Exosomes). Biochem. Soc. Trans. 2013, 41, 245–251.
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.M.; et al. Melanoma Exosomes Educate Bone Marrow Progenitor Cells toward a Pro-Metastatic Phenotype through MET. Nat. Med. 2012, 18, 883–891.
- Smyth, T.J.; Redzic, J.S.; Graner, M.W.; Anchordoquy, T.J. Examination of the Specificity of Tumor Cell Derived Exosomes with Tumor Cells in Vitro. Biochim. Biophys. Acta—Biomembr. 2014, 1838, 2954–2965.
- Rajagopal, C.; Harikumar, K.B. The Origin and Functions of Exosomes in Cancer. Front. Oncol. 2018, 8, 66.
- Santangelo, A.; Tamanini, A.; Cabrini, G.; Dechecchi, M.C. Circulating MicroRNAs as Emerging Non-Invasive Biomarkers for Gliomas. Ann. Transl. Med. 2017, 5, 1–8.
- Katakowski, M.; Chopp, M. Exosomes as Tools to Suppress Primary Brain Tumor. Cell. Mol. Neurobiol. 2016, 36, 343–352.
- Vaidya, M.; Sugaya, K. DNA Associated with Circulating Exosomes as a Biomarker for Glioma. Genes 2020, 11, 1276.
- Jafari, D.; Shajari, S.; Jafari, R.; Mardi, N.; Gomari, H.; Ganji, F.; Forouzandeh Moghadam, M.; Samadikuchaksaraei, A. Designer Exosomes: A New Platform for Biotechnology Therapeutics. BioDrugs 2020, 34, 567–586.
This entry is offline, you can click here to edit this entry!
