New and emerging plant diseases are caused by different pathogens including viruses that often cause significant crop losses. Badnaviruses are pararetroviruses that contain a single molecule of ds DNA genome of 7 to 9 kb in size and infect a large number of economically important crops such as banana and plantains, black pepper, cacao, citrus, grapevine, pineapple, sugarcane, sweet potato, taro, and yam, causing significant yield losses. Many of the species in the genus have a restricted host range and several of them are known to infect a single crop. Combined infections of different virus species and strains offer conditions that favor the development of new strains via recombination, especially in vegetatively propagated crops. The primary spread of badnaviruses is through vegetative propagating materials while for the secondary spread, they depend on insects such as mealybugs and aphids. Disease emerges as a consequence of the interactions between host and pathogens under favorable environmental conditions. The viral genome of the pararetroviruses is known to be integrated into the chromosome of the host and a few plants with integrants when subjected to different kinds of abiotic stress will give rise to episomal forms of the virus and cause disease.
- pararetrovirus
- reverse transcribing virus
- endogenous virus
- episomal virus
1. Introduction
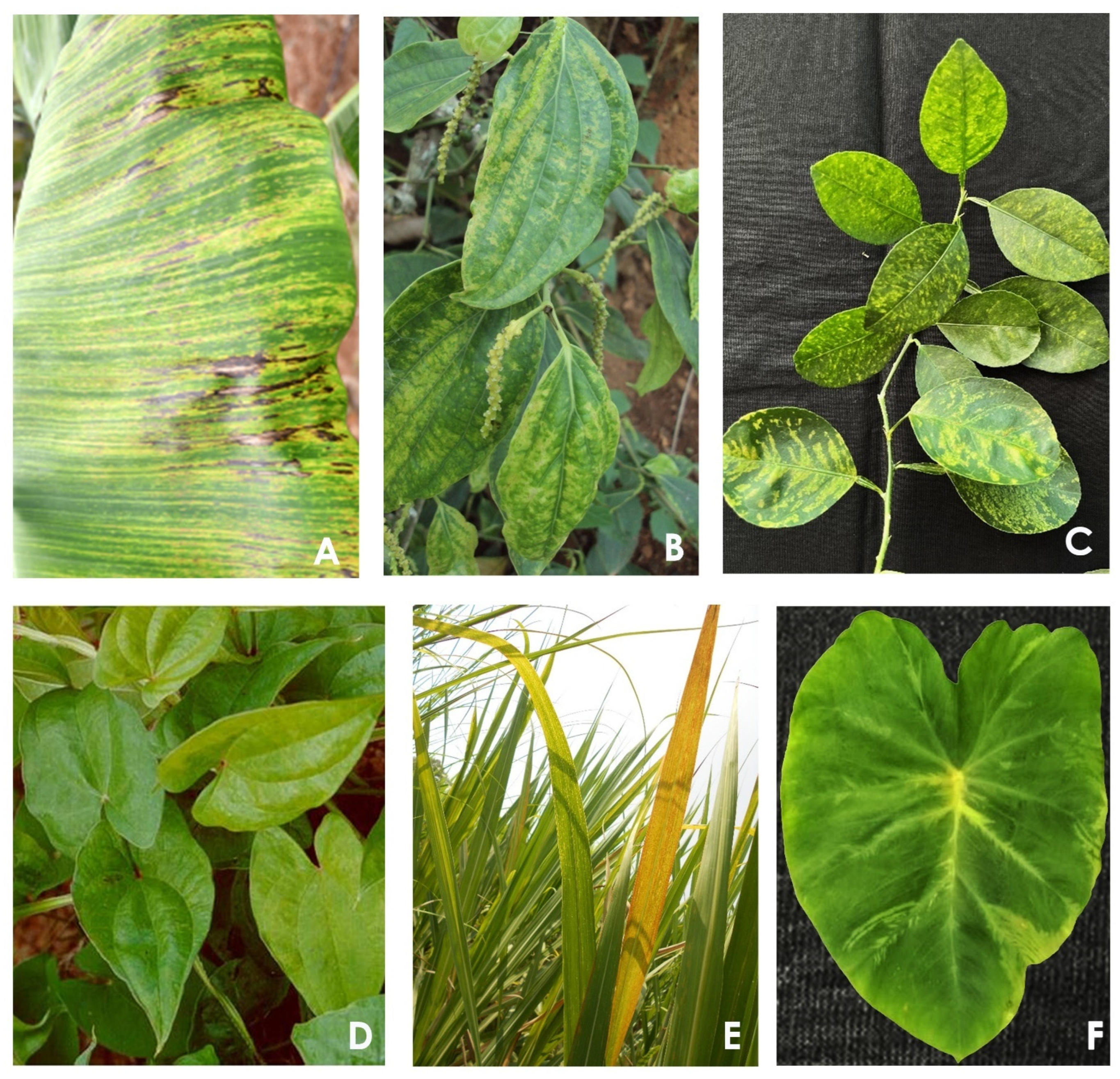
2. Emerging Disease Problems in Plants
2.1. Banana
2.2. Black Pepper
2.3. Citrus
2.4. Cacao
2.5. Dioscorea
2.6. Grapevine
2.7. Pineapple
2.8. Sugarcane
2.9. Sweet Potato
2.10. Taro
2.11. Badnaviruses Infecting Other Plants
3. Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/pathogens12020245
References
- King, A.M.Q.; Adams, M.J.; Lefkowitz, E.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2012; 1463p.
- International Committee on Taxonomy of Viruses (ICTV). Virus Taxonomy: 2021 Release. Available online: http://www.ictvonline.org/virusTaxonomy.asp (accessed on 28 November 2022).
- Chabannes, M.; Iskra-Caruana, M.-L. Endogenous pararetroviruses—A reservoir of virus infection in plants. Curr. Opin. Virol. 2013, 3, 615–620.
- Geering, A.D.W.; Hull, R. Family Caulimoviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: Boston, MA, USA, 2012; pp. 429–443.
- Bhat, A.I.; Hohn, T.; Selvarajan, R. Badnaviruses: The current global scenario. Viruses 2016, 8, 177.
- Borah, B.K.; Sharma, S.; Kant, R.; Anthony-Johnson, A.M.; Saigopal, D.V.R.; Dasgupta, I. Bacilliform DNA-containing plant viruses in the tropics: Commonalities within a genetically diverse group. Mol. Plant Pathol. 2013, 14, 759–771.
- Hohn, T.; Richert-Poggeler, K.R.; Harper, G.; Schawarzacher, T.; Teo, C.; Teycheney, P.-Y.; Iskra-Caruana, M.-L.; Hull, R. Evolution of integrated plant viruses. In Plant Virus Evolution; Roosinck, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 54–76.
- Staginnus, C.; Iskra-Caruana, M.; Lockhart, B.; Hohn, T.; Richert-Pöggeler, K.R. Suggestions for a nomenclature of endogenous pararetroviral sequences in plants. Arch. Virol. 2009, 154, 1189–1193.
- Dallot, S.; Accuna, P.; Rivera, C.; Ramirez, P.; Cote, F.; Lockhart, B.E.L.; Caruana, M.-L. Evidence that the proliferation stage of micropropagation procedure is determinant in the expression of Banana streak virus integrated into the genome of the FHIA21 hybrid (Musa AAAB). Arch. Virol. 2001, 146, 2179–2190.
- Lheureux, F.; Carreel, F.; Jenny, C.; Lockhart, B.E.L.; Iskra-Caruana, M.-L. Identification of genetic markers linked to banana streak disease expression in inter-specific Musa hybrids. Theor. Appl. Genet. 2003, 106, 594–598.
- Côte, F.X.; Galzi, S.; Folliot, M.; Lamagnère, Y.; Teycheney, P.-Y.; Iskra-Caruana, M.-L. Micropropagation by tissue culture triggers differential expression of infectious endogenous Banana streak virus sequences (eBSV) present in the B genome of natural and synthetic interspecific banana plantains. Mol. Plant Pathol. 2010, 11, 137–144.
- Dahal, G.; Hughes, J.D.A.; Thottappilly, G.; Lockhart, B.E.L. Effect of temperature on symptom expression and reliability of banana streak badnavirus detection in naturally infected plantain and banana (Musa spp.). Plant Dis. 1998, 82, 16–21.
- Dahal, G.; Ortiz, R.; Tenkouano, A.; Hughes, J.D.A.; Thottappilly, G.; Vuylsteke, D.; Lockhart, B.E.L. Relationship between natural occurrence of Banana streak badnavirus and symptom expression, relative concentration of viral antigen, and yield characteristics of some micropropagated Musa spp. Plant Pathol. 2000, 49, 68–79.
- Daniells, J.W.; Geering, A.D.W.; Bryde, N.J.; Thomas, J.E. The effect of Banana streak virus on the growth and yield of dessert bananas in tropical Australia. Ann. Appl. Biol. 2001, 139, 51–60.
- Lockhart, B.E.L. Purification and serology of a bacilliform virus associated with banana streak disease. Phytopathology 1986, 76, 995–999.
- Lockhart, B.E.L.; Kirtisak, K.A.; Jones, P.; Padmini, D.D.; Olszieewski, N.E.; Lockhart, N.; Nuarchan, D.; Sangalang, J. Identification of Piper yellow mottle virus, a mealy bug transmited badnavirus infecting Piper spp. in south East Asia. Eur. J. Plant Pathol. 1997, 103, 303–311.
- De Silva, D.P.; Jones, P.P.; Shaw, M.W. Identification and transmission of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum) in Sri Lanka. Plant Pathol. 2002, 51, 537–545.
- Bhat, A.I.; Devasahayam, S.; Sarma, Y.R.; Pant, R.P. Association of a Badnavirus in black pepper (Piper nigrum L.) transmitted by mealybug (Ferrisia virgata) in India. Curr. Sci. 2003, 84, 1547–1550.
- Che, H.; Cao, X.; Liu, P. Occurrence and characterization of virus species associated with black pepper (piper nigrum l.) virus diseases in Hainan province, China. J. Phytopathol. 2020, 169, 247–252.
- Bhat, A.I.; Biju, C.N.; Srinivasan, V.; Ankegowda, S.J.; Krishnamurthy, K.S. Current status of viral diseases affecting black pepper and cardamom. J. Spices Aromat. Crops 2018, 27, 1–16.
- Ahamedemujtaba, V.; Atheena, P.V.; Bhat, A.I.; Krishnamurthy, K.S.; Srinivasan, V. Symptoms of piper yellow mottle virus in black pepper as influenced by temperature and relative humidity. Virus Dis. 2021, 32, 305–313.
- Ahlawat, Y.S.; Pant, R.P.; Lockhart, B.E.L.; Srivastava, M.; Chakraborty, N.K.; Varma, A. Association of a badnavirus with citrus mosaic disease in India. Plant Dis. 1996, 80, 590–592.
- Huang, Q.; Hartung, J.S. Cloning and sequence analysis of an infectious clone of Citrus yellow mosaic virus that can infect sweet orange via Agrobacterium-mediated inoculation. J. Gen. Virol. 2001, 82, 2549–2558.
- Thresh, M.J. The origin and epidemiology of some important plant virus diseases. Appl. Biol. 1980, 5, 1–65.
- Ramos-Sobrinho, R.; Kouakou, K.; Bi, A.B.; Keith, C.V.; Diby, L.; Kouame, C.; Aka, R.A.; Marelli, J.-P.; Brown, J.K. Molecular detection of cacao swollen shoot badnavirus species by amplification with four PCR primer pairs, and evidence that Cacao swollen shoot Togo B virus-like isolates are highly prevalent in Côte d’Ivoire. Eur. J. Plant Pathol. 2021, 159, 941–947.
- Kandito, A.; Hartono, S.; Trisyono, Y.A.; Somowiyarjo, S. First report of Cacao mild mosaic virus associated with cacao mosaic disease in Indonesia. New Dis. Rep. 2022, 45, e12071.
- Muller, E.; Ravel, S.; Agret, C.; Abrokwah, F.; Dzahini-Obiatey, H.; Galyuon, I.; Kouakou, K.; Jeyaseelan, E.C.; Allainguillaume, J.; Wetten, A. Next generation sequencing elucidates cacao badnavirus diversity and reveals the existence of more than ten viral species. Virus Res. 2018, 244, 235–251.
- Chingandu, N.; Zia-ur-rehman, M.; Sreenivasan, T.N.; Surujdeo-Maharaj, S.; Umaharan, P.; Gutierrez, O.A.; Brown, J.K. Molecular characterization of previously elusive badnaviruses associated with symptomatic cacao in the New World. Arch. Virol. 2017, 162, 1363–1371.
- Ullah, I.; Daymond, A.J.; Hadley, P.; End, M.J.; Umaharan, P.; Dunwell, J.M. Identification of Cacao Mild Mosaic Virus (CaMMV) and Cacao Yellow Vein-Banding Virus (CYVBV) in Cocoa (Theobroma cacao) Germplasm. Viruses 2021, 13, 2152.
- Posnette, A.F. Virus diseases of cacao in West Africa VII: Virus transmission by different vector species. Ann. Appl. Biol. 1950, 37, 378–384.
- Marelli, J.P.; Guest, D.I.; Bailey, B.A.; Evans, H.C.; Brown, J.K.; Junaid, M.; Barreto, R.W.; Lisboa, D.O.; Puig, A.S. Chocolate under threat from old and new cacao diseases. Phytopathology 2019, 109, 1331–1343.
- Posnette, A.F. Virus diseases of cacao in West Africa. 1. Cacao viruses 1A, 1B, 1C and 1D. Ann. Appl. Biol. 1947, 34, 388–402.
- Jacquot, E.; Hagen, L.S.; Michler, P.; Rohfritsch, O.; Stussi-Garaud, C.; Keller, M.; Jacquemond, M.; Yot, P. In situ localisation of cacao swollen shoot virus in agroinoculated Theobroma cacao. Arch. Virol. 1999, 144, 259–271.
- Roivainen, O. Transmission of cocoa viruses by mealy bugs (Homoptera: Pseudococcidae). J. Sci. Agric. Soc. 1976, 48, 433–453.
- Quainoo, A.K.; Wetten, A.C.; Allainguillaume, J. Transmission of cocoa swollen shoot virus by seeds. J. Virol. Methods 2008, 150, 45–49.
- Ameyaw, G.A.; Wetten, A.; Dzahini-Obiatey, H.; Domfeh, O.; Allainguillaume, J. Investigation on Cacao swollen shoot virus (CSSV) pollen transmission through cross-pollination. Plant Pathol. 2013, 62, 421–427.
- Turaki, A.A.; Bömer, M.; Silva, G.; Lava Kumar, P.; Seal, S.E. PCR-DGGE analysis: Unravelling complex mixtures of badnavirus sequences present in yam germplasm. Viruses 2017, 9, 181.
- Bömer, M.; Rathnayake, A.I.; Visendi, P.; Silva, G.; Seal, S.E. Complete genome sequence of a new member of the genus Badnavirus, Dioscorea bacilliform RT virus 3, reveals the first evidence of recombination in yam badnaviruses. Arch. Virol. 2018, 163, 533–538.
- Bömer, M.; Turaki, A.; Silva, G.; Kumar, P.; Seal, S. A sequence-independent strategy for amplification and characterization of episomal badnavirus sequences reveals three previously uncharacterized yam badnaviruses. Viruses 2016, 8, 188.
- Umber, M.; Gomez, R.; Gelabale, S.; Bonheur, L.; Pavis, C.; Teycheney, P. The genome sequence of Dioscorea bacilliform TR virus, a member of the genus Badnavirus infecting Dioscorea spp., sheds light on the possible function of endogenous Dioscorea bacilliform viruses. Arch. Virol. 2016, 162, 517–521.
- Sukal, A.; Kidanemariam, D.; Dale, J.; James, A.; Harding, R. Characterization of badnaviruses infecting Dioscorea spp. in the Pacific reveals two putative novel species and the first report of Dioscorea bacilliform RT virus 2. Virus Res. 2017, 238, 29–34.
- Diouf, M.B.; Festus, R.; Silva, G.; Guyader, S.; Umber, M.; Seal, S.; Teycheney, P.Y. Viruses of Yams (Dioscorea spp.): Current Gaps in Knowledge and Future Research Directions to Improve Disease Management. Viruses 2022, 14, 1884.
- Zhang, Y.; Singh, K.; Kaur, R.; Qiu, W. Association of a novel DNA virus with the grapevine vein-clearing and vine decline syndrome. Phytopathology 2011, 101, 1081–1090.
- Cieniewicz, E.J.; Qiu, W.; Saldarelli, P.; Fuchs, M. Believing is seeing: Lessons from emerging viruses in grapevine. J. Plant Pathol. 2020, 102, 619–632.
- Rumbos, I.C.; Avgelis, A.D. Roditis leaf discoloration—A new virus disease of grapevine: Symptomatology and transmission to indicators plants. Phytopathol. Z. 1989, 152, 274–278.
- Maliogka, V.I.; Olmos, A.; Pappi, P.G.; Lotos, L.; Efthimiou, K.; Grammatikaki, G.; Candresse, T.; Katis, N.I.; Avgeli, A.D. A novel grapevine badnavirus is associated with the Roditis leaf discoloration disease. Virus Res. 2015, 203, 47–55.
- Chiumenti, M.; Morelli, M.; Giampetruzzi, A.; Palmisano, F.; Savino, V.N.; La Notte, P.; Martelli, G.P.; Saldarelli, P. First report of grapevine Roditis leaf discoloration-associated virus in Italy. J. Plant Pathol. 2015, 97, 551.
- Uluba, S.; Serçe, Ç.; Altan, B.; Bolat, V.; Ayyaz, M.; Çifçi, O.; Önder, S.; Öztürk Gökçe, Z.N.; Maliogka, V.I. First Report of grapevine Roditis leaf discoloration-associated virus infecting grapevine (Vitis vinifera) in Turkey. Plant Dis. 2018, 2, 256.
- Bester, R.; Lotos, L.; Vermeulen, A.; Pietersen, G.; Maliogka, V.I.; Maree, H.J. Complete genome sequence of a grapevine Roditis leaf discoloration-associated virus (GRLDaV) variant from South Africa. Arch. Virol. 2021, 166, 2041–2044.
- Ekemen, M. Investigation of Etiology on Grapevine Roditis Leaf Discoloration-Associated Virus. Master’s Thesis, Nigde Omer Halisdemir University, Niğde, Turkey, 2021. Available online: http://acikerisim.ohu.edu.tr/xmlui/handle/11480/8548 (accessed on 12 January 2023).
- Jagunic, M.; De Stradis, A.; Preiner, D.; La Notte, P.; Al Rwahnih, M.; Almeida, R.P.P.; Voncina, D. Biology and ultrastructural characterization of Grapevine Badnavirus 1 and Grapevine Virus G. Viruses 2022, 14, 2695.
- Vončina, D.; Almeida, R.P.P. Screening of some Croatian autochthonous grapevine varieties reveals a multitude of viruses, including novel ones. Arch. Virol. 2018, 163, 2239–2243.
- Wakman, W.D.; Teakle, D.S.; Thomas, J.E.; Dietzgen, R.G. Presence of clostero-like virus and a bacilliform virus in pineapple plants in Queensland. Aust. J. Agric. Res. 1995, 46, 947–958.
- Thomson, K.G.; Dietzgen, R.G.; Thomas, J.E.; Teakle, D.S. Detection of pineapple bacilliform virus using the polymerase chain reaction. Ann. Appl. Biol. 1996, 129, 57–69.
- Gambley, C.F.; Geering, A.D.; Steele, V.; Thomas, J.E. Identification of viral and non-viral reverse transcribing elements in pineapple (Ananas comosus), including members of two new badnavirus species. Arch. Virol. 2008, 153, 1599–1604.
- Sether, D.M.; Melzer, M.J.; Borth, W.B.; Hu, J.S. Pineapple bacilliform CO virus: Diversity, detection, distribution, and transmission. Plant Dis. 2012, 96, 1798–1804.
- Liting, W.; Xiaolei, R.; Wenjin, S. Sequencing and analysis of the complete genomic sequence of pineapple bacilliform comosus virus. China Agric. Sci. 2010, 43, 1969–1976.
- Autrey, L.J.C.; Boolell, S.; Jones, P. Distribution of sugarcane bacilliform virus in various geographical regions. In Proceedings of the XXI Congress of the International Society of Sugar Cane Technologists, Bangkok, Thailand, 5–14 March 1992; Kasetsart University: Bangkok, Thailand, 1995.
- Rao, G.P.; Sharma, S.K.; Kumar, P.V. Sugarcane Bacilliform Viruses: Present Status. In Plant Viruses; CRC Press: Boca Raton, FL, USA, 2018; pp. 117–130.
- Ahmad, K.; Sun, S.R.; Chen, J.L.; Huang, M.T.; Fu, H.Y.; Gao, S.J. Presence of diverse sugarcane bacilliform viruses infecting sugarcane in China revealed by pairwise sequence comparisons and phylogenetic analysis. Plant Pathol. J. 2019, 35, 41–50.
- Lockhart, B.E.L.; Autrey, L.J.C. Occurrence in sugarcane of a bacilliform virus related serologically to banana streak virus. Plant Dis. 1988, 72, 230–233.
- Lockhart, B.E.L.; Irey, M.J.; Comstock, J.C. Sugarcane bacilliform virus, Sugarcane mild mosaic virus, and sugarcane yellow leaf syndrome. In Sugarcane Germplasm Conservation and Exchange; Croft, B.J., Piggin, C.T., Wallis, E.S., Hogarth, D.M., Eds.; Australian Centre for International Agricultural Research (ACIAR): Brisbane, Australia, 1996; pp. 108–112.
- Bouhida, M.; Lockhart, B.E.; Olszewski, N.E. An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. J. Gen. Virol. 1993, 74, 15–22.
- Geijskes, R.J.; Braithwaite, K.S.; Dale, J.L. Sequence analysis of an Australian isolate of sugarcane bacilliform badnavirus. Arch. Virol. 2002, 147, 2393–2404.
- Muller, E.; Dupuy, V.; Blondin, L.; Bauffe, F.; Daugrois, J.H.; Nathalie, L.; Iskra-Caruana, M.L. High molecular variability of sugarcane bacilliform viruses in Guadeloupe implying the existence of at least three new species. Virus Res. 2011, 160, 414–419.
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388, 1–7.
- Kashif, M.; Pietilä, S.; Artola, K.; Jones, R.A.C.; Tugume, A.K.; Mäkinen, V.; Valkonen, J.P.T. Detection of viruses in sweetpotato from Honduras and Guatemala augmented by deep-sequencing of small-RNAs. Plant Dis. 2012, 96, 1430–1437.
- Mbanzibwa, D.R.; Tugume, A.K.; Chiunga, E.; Mark, D.; Tairo, F.D. Small RNA deep sequencing-based detection and further evidence of DNA viruses infecting sweetpotato plants in Tanzania. Ann. Appl. Biol. 2014, 165, 329–339.
- Buko, D.; Spetz, C.; Hvoslef-Eide, T. Next generation sequencing as a method to verify virus elimination using heat treatment and meristem tip culture in the five most widely used sweet potato varieties in Ethiopia. Afr. J. Biotechnol. 2020, 19, 458–463.
- Kreuze, J.F.; Perez, A.; Gargurevich, M.G.; Cuellar, W.J. Badnaviruses of sweet potato: Symptomless coinhabitants on a global scale. Front Plant Sci. 2020, 11, 313.
- Devitt, L.; Ebenebe, A.; Gregory, H.; Harding, R.; Hunter, D.; Macanawai, A. Investigations into the seed and mealybug transmission of Taro bacilliform virus. Aust. Plant Pathol. 2005, 34, 73–76.
- Yang, I.C.; Hafner, G.J.; Dale, J.L.; Harding, R.M. Genomic characterisation of taro bacilliform virus. Arch. Virol. 2003, 148, 937–949.
- Yang, I.C.; Hafner, G.J.; Revill, P.A.; Dale, J.L.; Harding, R.M. Sequence diversity of South Pacific isolates of Taro bacilliform virus and the development of a PCR-based diagnostic test. Arch. Virol. 2003, 148, 1957–1966.
- Stace-Smith, R.; Jones, A.T. Virus Diseases of Small Fruits; Converse, R.H., Ed.; United States Department of Agriculture: Washington, DC, USA, 1987; Section 4; p. 175.
- Marais, A.; Murolo, S.; Faure, C.; Brans, Y.; Larue, C.; Maclot, F.; Massart, S.; Chiumenti, M.; Minafra, A.; Romanazzi, G.; et al. Sixty Years from the First Disease Description, a Novel Badnavirus Associated with Chestnut Mosaic Disease. Phytopathology 2021, 111, 1051–1058.
- Rumbou, A.; Candresse, T.; Marais, A.; Theil, S.; Langer, J.; Jalkanen, R.; Büttner, C. A novel badnavirus discovered from Betula sp. affected by birch leaf-roll disease. PLoS ONE 2018, 13, e0193888.
- Wang, Y.; Cheng, X.; Wu, X.; Wang, A.; Wu, X. Characterization of complete genome and small RNA profile of Pagoda yellow mosaic associated virus, a novel Badnavirus in China. Virus Res. 2014, 188, 103–108.
- Chiumenti, M.; Morelli, M.; Elbeaino, T.; Stavolone, L.; De Stradis, A.; Digiaro, M.; Minafra, A.; Martelli, G.P. Sequencing an unconventional virus genome: The mulberry badnavirus 1 case. J. Plant Pathol. 2014, 96, 36–37.
- Alvarez-Quinto, R.A.; Lockhart, B.E.L.; Moreno-Martinez, J.M.; Olszewski, N.E. Complete genome sequence of aglaonema bacilliform virus (ABV). Arch. Virol. 2020, 165, 237–239.
- Uke, A.; Pinili, M.S.; Natsuaki, K.T.; Geering, A.D.W. Complete genome sequence of aucuba ringspot virus. Arch. Virol. 2021, 166, 1227–1230.
- Tsai, C.H.; Su, H.J.; Wu, M.-L.; Feng, Y.C.; Hung, T.H. Identification and detection of Bougainvillea spectabilis chlorotic vein-banding virus in different bougainvillea cultivars in Taiwan. Ann. Appl. Biol. 2008, 53, 187–193.
- Baranwal, V.K.; Meenakshi, A.; Singh, J. First report of two distinct badnaviruses associated with Bougainvillea spectabilis in India. J. Gen. Plant Pathol. 2010, 76, 236–239.
- Yusop, M.S.M.; Ersoy, R.; Akbar, M.A.; Saad, M.F.M.; Goh, H.H.; Baharum, S.N.; Bunawan, H. First report of Bougainvillea chlorotic vein-banding virus causing Chlorosis and vein banding of Bougainvillea spectabilis in Malaysia. Plant Dis. 2019, 103, 2974.
- Naito, F.Y.B.; De Nazaré Almeida dos Reis, L.; Batista, J.G.; Nery, F.M.B.; Rossato, M.; Melo, F.L.; De Cássia Pereira-Carvalho, R. Complete genome sequence of Bougainvillea chlorotic vein banding virus in Bougainvillea spectabilis from Brazil. Trop. Plant Pathol. 2020, 45, 159–162.
- Li, R.; Zheng, L.; Cao, M.; Wu, L.; Normandy, P.; Liu, H. First identification and molecular characterization of a new badnavirus infecting camellia. Arch. Virol. 2020, 165, 2115–2118.
- Wijayasekara, D.; Hoyt, P.; Gimondo, A.; Dunn, B.; Thapa, A.; Jones, H.; Verchot, J. Molecular characterization of two badnavirus genomes associated with Canna yellow mottle disease. Virus Res. 2018, 2, 19–24.
- Zhang, J.; Dey, K.K.; Lin, B.; Borth, W.B.; Melzer, M.J.; Sether, D.; Wang, Y.; Wang, I.C.; Shen, H.; Pu, X.; et al. Characterization of Canna yellow mottle virus in a New Host, Alpinia purpurata, in Hawaii. Phytopathology 2017, 107, 791–799.
- Lim, S.; Park, J.M.; Kwon, S.Y.; Cho, H.S.; Kim, H.S.; Lee, S.H.; Moon, J.S. Complete genome sequence of a tentative new member of the genus Badnavirus identified in Codonopsis lanceolata. Arch. Virol. 2019, 164, 1733–1737.
- Su, L.; Gao, S.; Huang, Y.; Ji, C.; Wang, D.; Ma, Y.; Fang, R.; Chen, X. Complete genomic sequence of Dracaena mottle virus, a distinct badnavirus. Virus Genes 2007, 35, 423–430.
- Lan, P.X.; Tian, T.Y.; Pu, L.L.; Rao, W.L.; Li, F.; Li, R.H. Characterization and detection of a new badnavirus infecting Epiphyllum spp. Arch. Virol. 2019, 164, 1837–1841.
- Bester, R.; Burger, J.T.; Maree, H.J. Genomic characterisation of a newly identified badnavirus infecting ivy (Hedera helix). Arch. Virol. 2020, 165, 1511–1514.
- Yang, Z.; Nicotaisen, M.; Olszewski, N.E.; Lockhart, B.E. Sequencing, improved detection, and a novel form of Kalanchoë top-spotting virus. Plant Dis. 2005, 89, 298–302.
- Diaz-Lara, A.; Mosier, N.J.; Keller, K.E.; Martin, R.R. A variant of Rubus yellow net virus with altered genomic organization. Virus Genes 2015, 50, 104–110.
- Alvarez-Quinto, R.A.; Serrano, J.; Olszewski, N.; Grinstead, S.; Mollov, D.; Lockhart, B.E.L. Complete genome sequences of two isolates of spiraea yellow leafspot virus (genus Badnavirus) from Spiraea x bumalda ‘Anthony Waterer’. Arch. Virol. 2022, 167, 631–634.
- Li, Y.; Deng, C.; Qiao, Y.; Zhao, X.; Zhou, Q. Characterization of a new badnavirus from Wisteria sinensis. Arch. Virol. 2017, 162, 2125–2129.
- Mehrvar, M.; Moradi, Z.; Al-Jaberi, M.S. First report of Wisteria badnavirus 1 infecting Wisteria sinensisin Iran. New Dis. Rep. 2022, 46, e12133.
- Alvarez-Quinto, R.A.; Lockhart, B.E.L.; Fetzer, J.L.; Olszewski, N.E. Genomic characterization of cycad leaf necrosis virus, the first badnavirus identified in a gymnosperm. Arch Virol. 2020, 165, 1671–1673.
- Shahid, M.S.; Aboughanem-Sabanadzovic, N.; Sabanadzovic, S.; Tzanetakis, I.E. Genomic Characterization and Population Structure of a Badnavirus Infecting Blackberry. Plant Dis. 2017, 101, 110–115.
- Laney, A.G.; Hassan, M.; Tzanetakis, I.E. An integrated badnavirus is prevalent in fig germplasm. Phytopathology 2012, 102, 1182–1189.
- Alishiri, A.; Rakhshandehroo, F.; Jouzani, G.S.; Shams-bakhsh, M. Exploring the genetic diversity and molecular evolution of fig badnavirus-1 from Iran. J. Plant Pathol. 2018, 100, 287–299.
- Xu, D.; Mock, R.; Kinard, G.; Li, R. Molecular analysis of the complete genomic sequences of four isolates of Gooseberry vein banding associated virus. Virus Genes 2011, 43, 130–137.
- Petrzik, K.; Přibylová, J.; Spak, J. Molecular analysis of gooseberry vein banding associated virus. Acta Virol. 2012, 56, 119–124.
- Zuļģe, N.; Stalažs, A.; Moročko-Bičevska, I.; Namniece, S.; Drevinska, K.; Konavko, D. Gooseberry vein banding associated virus on Ribes in Latvia: Occurrence, host plants and vectors. Plant Pathol. 2022, 71, 1910–1920.
- Du, K.; Liu, S.; Chen, Z.; Fan, Z.; Wang, H.; Tian, G.; Zhou, T. Full genome sequence of jujube mosaic-associated virus, a new member of the family Caulimoviridae. Arch. Virol. 2017, 162, 3221–3224.
- Liu, B.; Zhang, G.; Song, D.; Wang, Q.; Li, H.; Gu, A.; Bai, J. Complete genome sequence of a novel virus belonging to the genus Badnavirus in jujube (Ziziphus jujuba Mill.) in China. Arch. Virol. 2022, 167, 1885–1888.
- Alvarez-Quinto, R.A.; Lockhart, B.E.L.; Olszewski, N. Complete genome sequence of a previously undescribed badnavirus occurring in Polyscias fruticosa L. (Ming aralia). Arch Virol. 2019, 164, 2371–2374.
- Xu, M.; Zhang, S.; Xuan, Z.; Wu, J.; Dong, P.; Zhou, Y.; Li, R.; Cao, M. Molecular characterization of a new badnavirus infecting green Sichuan pepper (Zanthoxylum schinifolium). Arch. Virol. 2019, 164, 2613–2616.
- Lee, Y.J.; Kwak, H.R.; Lee, Y.K.; Kim, M.K.; Choi, H.S.; Seo, J.K. Complete genome sequence of yacon necrotic mottle virus, a novel putative member of the genus Badnavirus. Arch. Virol. 2015, 160, 1139–1142.
- Medberry, S.L.; Lockhart, B.E.L.; Olszewski, N.E. The Commelinayellow m ottlevirus promoter is a strong promoter in vascular and reproductive tissues. Plant Cell 1992, 4, 185–192.
