Hepatitis C Virus (HCV) is an enveloped virus with an RNA genome of positive polarity. HCV replicates mainly in the liver and can cause liver disease, cirrhosis and cancer. Translation of the HCV RNA genome is regulated by the Internal Ribosome Entry Site (IRES) and other cis-elements in the viral RNA genome. The viral RNA usurps cellular ribosomes using a variety of viral RNA elements as well as by recruiting cellular RNA binding proteins. Here, we give a comprehensive overview over the determinants involved in the regulation of HCV translation.
- HCV
- Hepacivirus
- Internal Ribosome Entry Site
- IRES
- Initiation
- Ribosome
- 40S
- eIF3
- ITAF
- stress response
1. Introduction
Hepatitis C virus (HCV) is an enveloped positive strand RNA virus that preferentially replicates in the liver [1], and it is classified in the genus Hepacivirus in the family Flaviviridae. Worldwide, about 71 million people are infected with HCV [2]. The infection is usually noticed only when coincidentally diagnosed by routine testing, for example, during hospitalization, or when the liver disease becomes acute. In the latter case, liver damage by virus replication and the resulting immune responses can lead to impaired bilirubin conjugation in the liver, and unconjugated bilirubin deposits can then be noticed as a yellowish color (called jaundice), often first in the sclera in the eyes and when more severe also in the skin. An acute infection can result in severe liver damage, in rare cases even resulting in death [3][4]. However, most HCV infections remain inapparent [5][6], and the virus infection can become chronic in about 60% to 70 % of all infections [7], often without being noticed. Chronic infection can, in the long run, result in liver cirrhosis and liver cancer (hepatocellular carcinoma, HCC) [8][9][10], while a metabolic reprogramming of the infected cells according to the “Warburg effect” like in cancer cells can be observed only a few days after the onset of HCV replication [11]. Moreover, inapparent replication of the virus usually results in unnoticed spread of the virus to other individuals, a fact that is a major challenge for surveillance, health care, and treatment [12]. Meanwhile, very effective treatment regimens using direct acting antivirals (DAAs) are available, although they are still very expensive [13][14]. Although the error rate of the viral replicase is high and can, in principle, easily give rise to resistance mutations, the conserved nature of the replicase active center and the often occurring reduced fitness of mutants result in the rare appearance of resistance mutations against nucleoside inhibitors such as sofosbuvir [15][16]. An effective vaccination is not yet available, also partially due to the high variability of the viral RNA genome. Thus, further research on HCV is urgently required to combat HCV infections, and despite much progress in the understanding of HCV replication, the molecular mechanisms of HCV replication are still far from being completely understood [12].
2. Hepatitis C virus (HCV)
The HCV RNA genome is about 9600 nucleotides (nts) long and codes for one long polyprotein that is co- and post-translationally processed into the mature gene products [17][18][19]. Unlike most cellular mRNAs, the 5′-end of the HCV genomic RNA has no cap nucleotide attached which would govern efficient cap-dependent translation initiation [20][21][22]. Instead, HCV translation is mediated by virtue of an internal ribosome entry site (IRES) [23][24][25][26][27], which is located largely in the 5′UTR but also slightly spans into the coding region (Figure 1). While the resulting low efficient translation coincides with the “undercover” strategy of HCV replication that often leads to chronic infection and further unnoticed spread of the virus to uninfected individuals, the use of such IRES elements has two more big advantages. The first advantage is that in particular the very ends of the RNA genome do not need to serve functions in translation control such as in capped and polyadenylated cellular mRNAs. Instead, RNA signals that are involved in genome replication can be directly placed at the very genome ends [28][18][29]. The second benefit of cap-independent translation is that the virus escapes antiviral countermeasures of the cell in terms of the downregulation of cap-dependent translation, which is largely conferred by phosphorylation of eIF2 and the resulting inhibition of cap-dependent translation initiation [30].
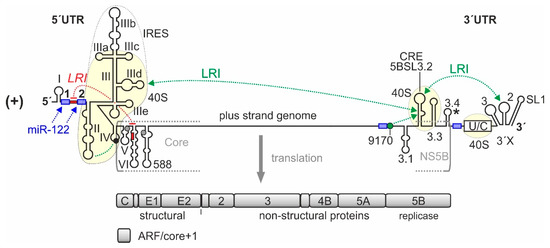
Figure 1.
-Elements in the hepatitis C virus (HCV) RNA genome that are involved in translation regulation. The HCV plus strand RNA genome. The internal ribosome entry site (IRES) in the HCV 5′-untranslated region (5′UTR), the entire 3′UTR and the c
-acting replication element (CRE) in the NS5B coding region are involved in translation regulation [23][28][31]. Those regions of the 5′UTR, 3′UTR, and CRE that bind to the ribosomal 40S subunit are underlayed in light yellow. Stem-loops (SLs) in the 5′UTR are numbered by roman numerals. The region of the IRES is surrounded by a dotted line. The IRES includes SLs II–IV of the 5′UTR but spans into the core protein coding region. The canonical AUG start codon in SL IV of the 5′UTR (black circle) gives rise to translation of the polyprotein which is cleaved to yield structural proteins and non-structural (NS-) proteins, including the viral replicase NS5B. The 3′UTR contains the variable region, a poly(U/C) tract (U/C), and the so-called 3′X region including SLs 1, 2, and 3. The stem-loop 5BSL3.2 in the 3′-region of the NS5B coding region is the CRE, flanked by upstream stem-loop 5BSL3.1 and downstream 5BSL3.3. The polyprotein stop codon is located in the stem-loop 5BSL3.4 (asterisk). Some other start codons which give rise to the alternative reading frame (ARF) in the core+1 reading frame [32][33][34] are shown in grey. Positively and negatively acting long-range RNA–RNA interactions (LRIs) are shown in green or red, respectively, with the sequence “9170” shown as green circle. Selected binding sites for microRNA-122 (miR-122) are shown in blue.
In this entry, we focus on the sequences, cellular factors, and molecular mechanisms involved in the regulation of translation by the HCV IRES. Thereby, we touch the functions of miR-122 specifically only with regard to translation regulation.
