Bladder cancer is one of the most common malignancies of the urinary tract and can be divided into non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC). Emerging evidence demonstrates that long noncoding RNAs play a crucial role in the carcinogenesis and progression of bladder cancer. Long intergenic noncoding RNAs (lincRNAs) are a subgroup of long noncoding RNAs (lncRNAs) that do not overlap protein-coding genes. Small nucleolar RNAs (snoRNAs) are a class of noncoding RNAs (ncRNAs) that mainly exist in the nucleolus, are approximately 60–300 nucleotides in length, and are hosted inside the introns of genes. Small nucleolar RNA host genes (SNHGs) have been associated with the origin and development of bladder cancer.
- bladder cancer
- small nucleolar RNA
- long intergenic noncoding RNA
1. Introduction
2. Pathophysiology of BLCA
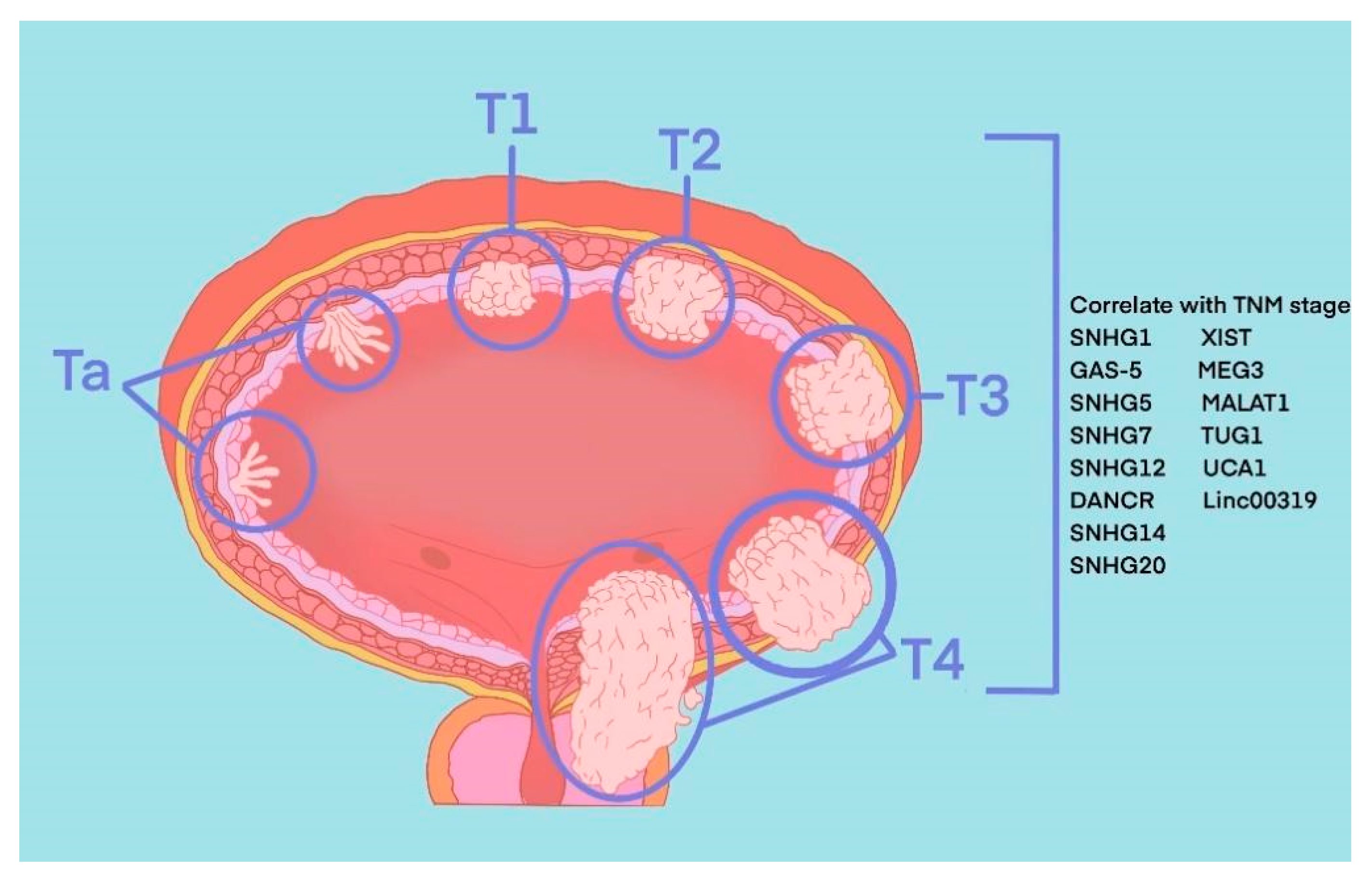
The most common pathological type of BLCA is bladder urothelial carcinoma. According to the depth of tumour invasion to bladder mucosa, BLCA can be divided into NMIBC and MIBC. Specifically, tumours at the Ta stage, T1 stage and carcinoma in situ (CIS) belong to NMIBC, and tumours at T3 and T4 stages belong to MIBC. These two types represent two periods of occurrence and development of BLCA, and their treatment methods and prognosis of them are quite different. TURBT and intravesical chemotherapy are the main treatment methods of the former, while the latter requires radical cystectomy or systemic treatment. Recently, in the published literature indicated 8 SNHGs (SNHG1, SNHG2, SNHG5, SNHG7, SNHG12-14 and SNHG20) and 6 lincRNAs (linc0001, linc00023, linc00047, linc00080, linc00178 and linc00319) that correlate with the TNM stage of BLCA (Figure 1).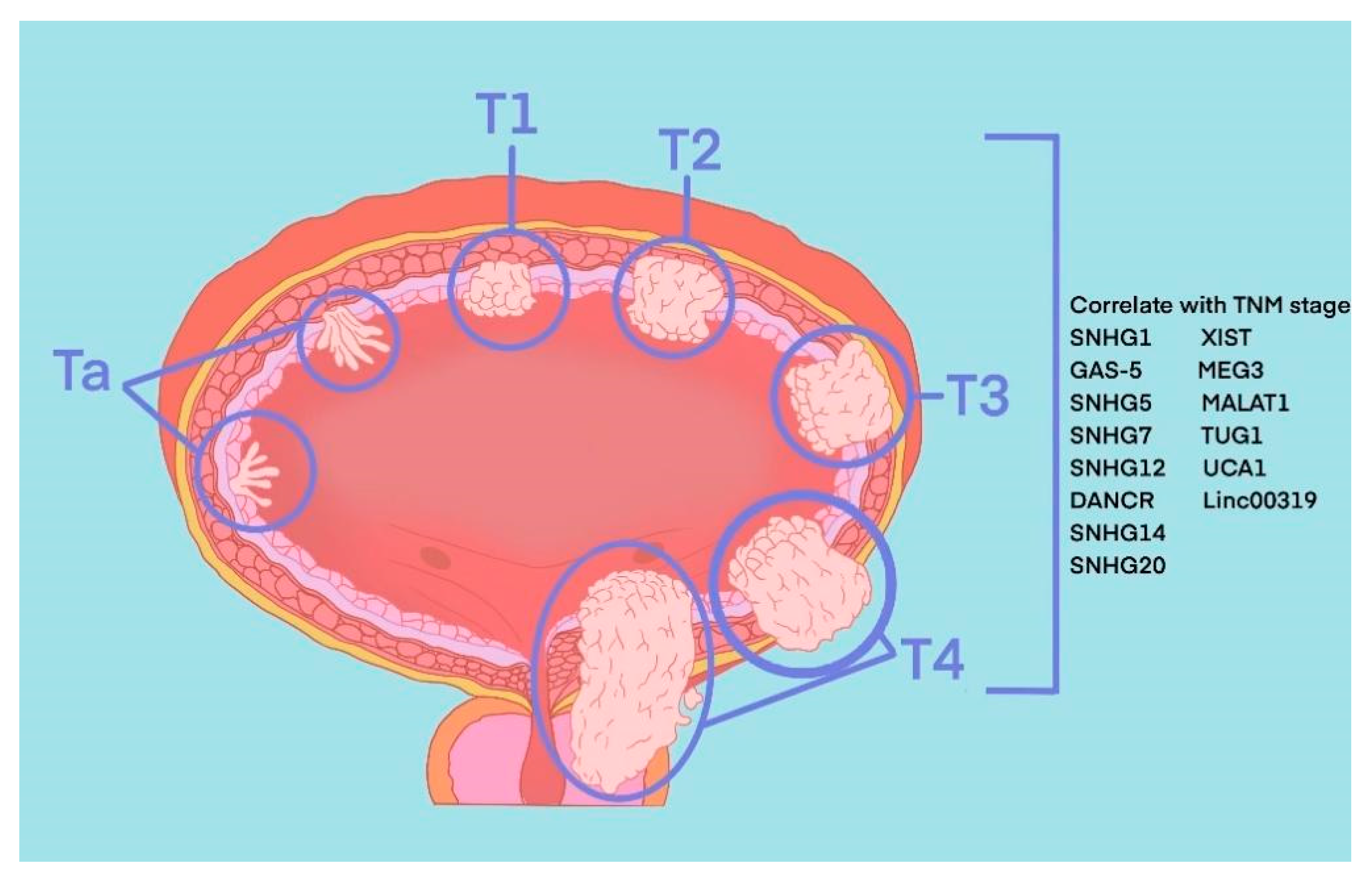
3. SnoRNA and lincRNA
4. The Role of snoRNA in BLCA
SNHG1, which is located at chromosome 11q12.3, is overexpressed in several types of tumours, including pancreatic, prostate, non-small-cell lung and BLCA [21,22,23,24][15][16][17][18]. It is widely considered an oncogene that promotes cancer proliferation, migration, invasion and tumorigenesis. SNHG1 can bind and coregulate with the PP2A catalytic subunit (PP2A-c) to promote c-Jun phosphorylation. Then, activated c-Jun increases matrix metalloproteinase 2 (MMP2) transcription, which induces cancer cell invasion and metastasis. Additionally, transcription of miR-34a was downregulated by SNHG1 via autophagy, thus maintaining MM2 mRNA stabilization [21][15].
SNHG2, which is also known as growth arrest-specific transcript 5 (GAS-5), is located at chromosome 1q25 and acts as a tumour suppressor in multiple cancers [26,27,28,29][19][20][21][22]. Studies have found that SNHG2 is expressed at low levels in BLCA cells and tissues and that the expression level of SNHG2 is markedly correlated with the clinical characteristics and prognosis of BLCA. High expression of SNHG2 promotes BLCA cell apoptosis and inhibits tumour proliferation. SNHG2 directly interacts with E2F transcription factor 4 (E2F4) and recruits it to the enhancer of the zeste homolog 2 (EZH2) promoter, thereby downregulating transcription of the EZH2 oncogene [26][19]. In the BLCA cell line HTB-9, SNHG2 sponges miR-21, thus promoting transcription of its downstream gene phosphatase and tensin homolog (PTEN), leading to the downregulation of antiapoptotic proteins and cell cycle-associated proteins, which suppresses proliferation and increases apoptosis of bladder cancer cells [27][20]. According to previous studies, downregulation of SNHG2 could also directly arrest the cell cycle at the S phase in a cyclin-dependent kinase 6 (CDK6)-dependent manner [28][21].
5. The Role of lincRNA in BLCA
Linc00001, also known as X-inactive specific transcript (XIST), is located on chromosome Xq13.2. It was first found in the process of X chromosome inactivation, which occurs at the early development stage of mammalian females, and was later shown to act as an oncogene and to be upregulated in multiple types of cancers, including BLCA [50][33]. Recent studies have revealed that XIST can regulate cancer cell progression by acting as a sponge of miRNAs and binding to proteins [51,52,53,54][34][35][36][37]. Specifically, the expression of XIST was elevated in BLCA tissues and cell lines, including T24, 253J, RT112 and HT-1376. XIST silencing induced the loss of proliferation, metastasis and stemness capability combined with the overexpression of miR-200c and miR-133a in vitro [53,54][36][37].
Linc00023 is located on chromosome 14q32.2 within the DLK-MEG3 locus and is also widely known as the maternally expressed 3 gene (MEG3). The deregulation of linc00023 was reported to be involved in the progression of various tumours, including meningioma, hepatocellular cancer, breast cancer and BLCA [55,56][38][39]. Similarly, MEG3 can also act as a ceRNA to regulate tumour cell proliferation and apoptosis. In bladder cancer, MEG3 was found to function as a ceRNA for PTEN by competitively binding miR-494, thus repressing the proliferation, migration and invasion and promoting the apoptosis of tumour cells [57][40].
Linc00047 is also known as metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and has a length of 8.5 kb. It is located on chromosome 11q13, is involved in the regulation of several molecular signalling pathways and has been shown to be a potential biomarker in bladder cancer, nasopharyngeal carcinoma and osteosarcoma [61,62][41][42]. The expression of MALAT1 was significantly elevated in BLCA tissues and cell lines and was positively correlated with advanced clinical stage and poor prognosis [63][43]. In addition, MALAT1 sponged miR-101-3p and suppressed the expression of VEGF-C in BLCA 5637 and EJ-M3 cell lines, enhancing cisplatin sensitivity. Furthermore, the knockdown of MALAT1 in cisplatin-resistant 5637 and EJ-M3 cells could reverse drug resistance [64][44].
Linc00080, also known as taurine-upregulated gene 1 (TUG1), is located on chromosome 22q12 and is almost twice as elevated in bladder tissues compared to adjacent normal tissues. It is broadly considered an oncogene in several tumours, such as colorectal cancer, oesophageal cancer, osteosarcoma and bladder cancer [66][45]. In BLCA, TUG1 is involved in tumour proliferation, metastasis and apoptosis, as well as radioresistance and chemotherapy resistance [67,68,69,70,71][46][47][48][49][50]. Silencing TUG1 in BLCA cell lines restrains the tendency of EMT by acting as a ceRNA of ZEB2 by sponging miR-145 and leading to radioresistance [67][46]. It also promotes the expression of high mobility group box 1 protein (HMGB1), thus enhancing bladder cancer radioresistance in vivo and in vitro, yet the mechanism needs further exploration [68][47].
Linc00178 is located on 19p13.12 and was first identified in bladder transitional cell carcinoma; thus, linc00178 is also known as urothelial cancer associated 1 (UCA1). Recent studies have verified the overexpression and oncogene functions of UCA1 in different tumours [72][51]. Specifically, UCA1 can serve as a sponge of miRNAs to regulate tumour growth, metastasis, drug resistance and mitochondrial functions. Li et al. found that knockdown of UCA1 in the BLCA cell line 5637 decreased mitochondrial DNA copy number by more than half with accompanying decreased ATP production. The results of a UCA1 overexpression experiment on the BLCA cell line UMUC2 were highly consistent with those of the knockdown experiments.
Linc00319 is a novel lncRNA located on chromosome 21q22.3 that has been implicated in the tumorigenesis and progression of cervical cancer, gastric cancer, osteosarcoma and laryngeal squamous cell carcinoma in a miRNA-dependent manner [77,78,79,80][52][53][54][55]. Moreover, linc00319 expression levels were remarkably higher in BLCA tissues than in adjacent normal tissues. Patients with higher linc00319 levels had higher clinical stages and lower recurrence-free survival rates [81][56].
Linc00355 is located on chromosome 13q21.31 and has been validated to function as a ceRNA by sponging miRNAs in lung squamous cell carcinoma, glioma and hepatocellular carcinoma [83,84,85][57][58][59]. In BLCA, linc00355 could act as a sponge of miR-424-5p to modulate high mobility group AT-hook 2 (HMGA2) expression, which could regulate the EMT-related proteins ZEB1, E-cadherin and vimentin and finally contribute to BLCA EMT and lung metastasis [86][60].
Linc00649 is located on 21q22.11 and is widely considered an oncogene in multiple tumours, such as lung squamous cell carcinoma, breast cancer and gastric cancer [94,95,96][61][62][63]. A recent study found that linc00649 is a basement membrane-related lncRNA and is correlated with clinical prognosis based on analysis of transcriptional and clinical data of bladder cancer from the TCGA, GEO and BM-BASE databases, revealing that linc00649 is a potential biomarker of BLCA. Additionally, a model containing eight lncRNAs, including linc00649, was constructed and used to accurately predict the prognosis of BLCA patients [97][64].
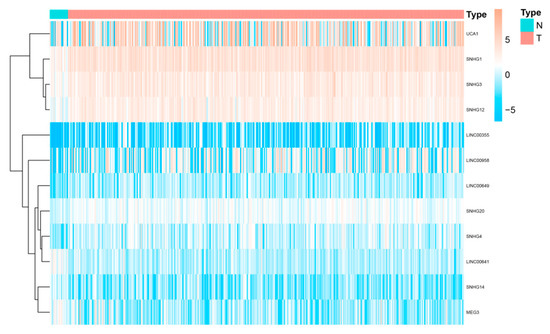
Linc00958 is located on 11p15.3 and was found to be substantially expressed in bladder cancer tissues compared to normal bladder epithelial tissues. Increasingly, studies have demonstrated that linc00958 is involved in the malignant progression of various cancers, such as hepatocellular carcinoma, colorectal cancer, osteosarcoma and endometrial cancer [100][65]. Anna et al. identified 72 BLCA tissues and 8 normal bladder epithelial tissues by RNA sequencing and selected five significantly dysregulated lncRNAs, including linc00958, for further analysis. They found that the knockdown of linc00958 led to a loss of cell mobility in vitro.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33.
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436.
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407.
- Palazzo, A.F.; Koonin, E.V. Functional Long Non-coding RNAs Evolve from Junk Transcripts. Cell 2020, 183, 1151–1161.
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: LncRNA localization and function. J. Cell Biol. 2021, 220, e20200904.
- Dsouza, V.L.; Adiga, D.; Sriharikrishnaa, S.; Suresh, P.S.; Chatterjee, A.; Kabekkodu, S.P. Small nucleolar RNA and its potential role in breast cancer—A comprehensive review. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188501.
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer 2012, 12, 84–88.
- Ding, Y.; Sun, Z.; Zhang, S.; Li, Y.; Han, X.; Xu, Q.; Zhou, L.; Xu, H.; Bai, Y.; Xu, C.; et al. Downregulation of snoRNA SNORA52 and Its Clinical Significance in Hepatocellular Carcinoma. Biomed. Res. Int. 2021, 2021, 7020637.
- Bao, H.-J.; Chen, X.; Liu, X.; Wu, W.; Li, Q.-H.; Xian, J.-Y.; Zhao, Y.; Chen, S. Box C/D snoRNA SNORD89 influences the occurrence and development of endometrial cancer through 2′-O-methylation modification of Bim. Cell Death Discov. 2022, 8, 309.
- Rinn, J.L.; Chang, H.Y. Long Noncoding RNAs: Molecular Modalities to Organismal Functions. Annu. Rev. Biochem. 2020, 89, 283–308.
- Martens-Uzunova, E.S.; Böttcher, R.; Croce, C.M.; Jenster, G.; Visakorpi, T.; Calin, G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014, 65, 1140–1151.
- Reichow, S.L.; Hamma, T.; Ferré-D'Amaré, A.R.; Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007, 35, 1452–1464.
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220.
- Zimta, A.-A.; Tigu, A.B.; Braicu, C.; Stefan, C.; Ionescu, C.; Berindan-Neagoe, I. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front. Oncol. 2020, 10, 389.
- Xu, J.; Yang, R.; Hua, X.; Huang, M.; Tian, Z.; Li, J.; Lam, H.Y.; Jiang, G.; Cohen, M.; Huang, C. lncRNA SNHG1 Promotes Basal Bladder Cancer Invasion via Interaction with PP2A Catalytic Subunit and Induction of Autophagy. Mol. Ther. Nucleic Acids 2020, 21, 354–366.
- Xu, J.; Yang, R.; Li, J.; Wang, L.; Cohen, M.; Simeone, D.M.; Costa, M.; Wu, X.-R. DNMT3A//Rac1 Is an Effector Pathway for to Drive Stem-Cell-like and Invasive Behaviors of Advanced Bladder Cancer Cells. Cancers 2022, 14, 4159.
- Xiang, W.; Lyu, L.; Huang, T.; Zheng, F.; Yuan, J.; Zhang, C.; Jiang, G. The long non-coding RNA SNHG1 promotes bladder cancer progression by interacting with miR-143-3p and EZH2. J. Cell. Mol. Med. 2020, 24, 11858–11873.
- Min, J.; Ma, J.; Wang, Q.; Yu, D. Long non-coding RNA SNHG1 promotes bladder cancer progression by upregulating EZH2 and repressing KLF2 transcription. Clinics 2022, 77, 100081.
- Wang, M.; Guo, C.; Wang, L.; Luo, G.; Huang, C.; Li, Y.; Liu, D.; Zeng, F.; Jiang, G.; Xiao, X. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018, 9, 238.
- Chen, D.; Guo, Y.; Chen, Y.; Guo, Q.; Chen, J.; Li, Y.; Zheng, Q.; Jiang, M.; Xi, M.; Cheng, L. LncRNA growth arrest-specific transcript 5 targets miR-21 gene and regulates bladder cancer cell proliferation and apoptosis through PTEN. Cancer Med. 2020, 9, 2846–2858.
- Liu, Z.; Wang, W.; Jiang, J.; Bao, E.; Xu, D.; Zeng, Y.; Tao, L.; Qiu, J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS ONE 2013, 8, e73991.
- Zhang, H.; Guo, Y.; Song, Y.; Shang, C. Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. Cancer Chemother. Pharmacol. 2017, 79, 49–55.
- Cao, Y.; Hu, Q.; Zhang, R.; Li, L.; Guo, M.; Wei, H.; Zhang, L.; Wang, J.; Li, C. Knockdown of Long Non-coding RNA SNGH3 by CRISPR-dCas9 Inhibits the Progression of Bladder Cancer. Front. Mol. Biosci. 2021, 8, 657145.
- Xie, J.; Ni, J.; Shi, H.; Wang, K.; Ma, X.; Li, W.; Peng, B. LncRNA SNHG3 enhances BMI1 mRNA stability by binding and regulating c-MYC: Implications for the carcinogenic role of SNHG3 in bladder cancer. Cancer Med. 2022.
- Dai, G.; Huang, C.; Yang, J.; Jin, L.; Fu, K.; Yuan, F.; Zhu, J.; Xue, B. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J. Cell. Mol. Med. 2020, 24, 9231–9243.
- Li, Y.-H.; Hu, Y.-Q.; Wang, S.-C.; Li, Y.; Chen, D.-M. LncRNA SNHG5: A new budding star in human cancers. Gene 2020, 749, 144724.
- Wang, C.; Tao, W.; Ni, S.; Chen, Q. Upregulation of lncRNA snoRNA host gene 6 regulates NUAK family SnF1-like kinase-1 expression by competitively binding microRNA-125b and interacting with Snail1/2 in bladder cancer. J. Cell. Biochem. 2019, 120, 357–367.
- Zhou, Y.; Tian, B.; Tang, J.; Wu, J.; Wang, H.; Wu, Z.; Li, X.; Yang, D.; Zhang, B.; Xiao, Y.; et al. SNHG7: A novel vital oncogenic lncRNA in human cancers. Biomed. Pharmacother. 2020, 124, 109921.
- Zhao, Q.; Gao, S.; Du, Q.; Liu, Y. Long non-coding RNA SNHG20 promotes bladder cancer via activating the Wnt/β-catenin signalling pathway. Int. J. Mol. Med. 2018, 42, 2839–2848.
- Wang, W.; Chen, S.; Song, X.; Gui, J.; Li, Y.; Li, M. ELK1/lncRNA-SNHG7/miR-2682-5p feedback loop enhances bladder cancer cell growth. Life Sci. 2020, 262, 118386.
- Kretz, M.; Webster, D.E.; Flockhart, R.J.; Lee, C.S.; Zehnder, A.; Lopez-Pajares, V.; Qu, K.; Zheng, G.X.Y.; Chow, J.; Kim, G.E.; et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012, 26, 338–343.
- Chen, Z.; Chen, X.; Xie, R.; Huang, M.; Dong, W.; Han, J.; Zhang, J.; Zhou, Q.; Li, H.; Huang, J.; et al. DANCR Promotes Metastasis and Proliferation in Bladder Cancer Cells by Enhancing IL-11-STAT3 Signaling and CCND1 Expression. Mol. Ther. 2019, 27, 326–341.
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647.
- Chen, D.; Chen, T.; Guo, Y.; Wang, C.; Dong, L.; Lu, C. Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo. Exp. Cell Res. 2020, 396, 112281.
- Hu, B.; Shi, G.; Li, Q.; Li, W.; Zhou, H. Long noncoding RNA XIST participates in bladder cancer by downregulating p53 via binding to TET1. J. Cell. Biochem. 2019, 120, 6330–6338.
- Xu, R.; Zhu, X.; Chen, F.; Huang, C.; Ai, K.; Wu, H.; Zhang, L.; Zhao, X. LncRNA XIST/miR-200c regulates the stemness properties and tumourigenicity of human bladder cancer stem cell-like cells. Cancer Cell Int. 2018, 18, 41.
- Zhou, K.; Yang, J.; Li, X.; Chen, W. Long non-coding RNA XIST promotes cell proliferation and migration through targeting miR-133a in bladder cancer. Exp. Ther. Med. 2019, 18, 3475–3483.
- Al-Rugeebah, A.; Alanazi, M.; Parine, N.R. MEG3: An Oncogenic Long Non-coding RNA in Different Cancers. Pathol. Oncol. Res. 2019, 25, 859–874.
- Hsieh, P.-F.; Yu, C.-C.; Chu, P.-M.; Hsieh, P.-L. Long Non-Coding RNA MEG3 in Cellular Stemness. Int. J. Mol. Sci. 2021, 22, 5348.
- Shan, G.; Tang, T.; Xia, Y.; Qian, H.-J. MEG3 interacted with miR-494 to repress bladder cancer progression through targeting PTEN. J. Cell. Physiol. 2020, 235, 1120–1128.
- Li, Z.-X.; Zhu, Q.-N.; Zhang, H.-B.; Hu, Y.; Wang, G.; Zhu, Y.-S. MALAT1: A potential biomarker in cancer. Cancer Manag. Res. 2018, 10, 6757–6768.
- Goyal, B.; Yadav, S.R.M.; Awasthee, N.; Gupta, S.; Kunnumakkara, A.B.; Gupta, S.C. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188502.
- Jiao, D.; Li, Z.; Zhu, M.; Wang, Y.; Wu, G.; Han, X. LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC). Am. J. Cancer Res. 2018, 8, 748–760.
- Liu, P.; Li, X.; Cui, Y.; Chen, J.; Li, C.; Li, Q.; Li, H.; Zhang, X.; Zu, X. LncRNA-MALAT1 mediates cisplatin resistance via miR-101-3p/VEGF-C pathway in bladder cancer. Acta Biochim. Biophys. Sin. 2019, 51, 1148–1157.
- Li, Z.; Shen, J.; Chan, M.T.V.; Wu, W.K.K. TUG1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016, 49, 471–475.
- Tan, J.; Qiu, K.; Li, M.; Liang, Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015, 589 Pt B, 3175–3181.
- Jiang, H.; Hu, X.; Zhang, H.; Li, W. Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat. Oncol. 2017, 12, 65.
- Yuan, J.-B.; Gu, L.; Chen, L.; Yin, Y.; Fan, B.-Y. Annexin A8 regulated by lncRNA-TUG1/miR-140-3p axis promotes bladder cancer progression and metastasis. Mol. Ther. Oncolytics 2021, 22, 36–51.
- Yu, G.; Zhou, H.; Yao, W.; Meng, L.; Lang, B. lncRNA TUG1 Promotes Cisplatin Resistance by Regulating CCND2 via Epigenetically Silencing miR-194-5p in Bladder Cancer. Mol. Ther. Nucleic Acids 2019, 16, 257–271.
- Liu, Q.; Liu, H.; Cheng, H.; Li, Y.; Li, X.; Zhu, C. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. Onco Targets Ther. 2017, 10, 2461–2471.
- Ghafouri-Fard, S.; Taheri, M. UCA1 long non-coding RNA: An update on its roles in malignant behavior of cancers. Biomed. Pharmacother. 2019, 120, 109459.
- Sun, F.; Yu, Z.; Wu, B.; Zhang, H.; Ruan, J. LINC00319 promotes osteosarcoma progression by regulating the miR-455-3p/NFIB axis. J. Gene Med. 2020, 22, e3248.
- Ma, Z.; Cai, Y.; Zhang, L.; Tian, C.; Lyu, L. LINC00319 Promotes Cervical Cancer Progression Via Targeting miR-147a/IGF1R Pathway. Cancer Biother. Radiopharm. 2020.
- Zou, J.; Wu, K.; Lin, C.; Jie, Z.-G. LINC00319 acts as a sponge to accelerate tumor growth and metastasis in gastric cancer by upregulating. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G10–G22.
- Yuan, L.; Tian, X.; Zhang, Y.; Huang, X.; Li, Q.; Li, W.; Li, S. LINC00319 promotes cancer stem cell-like properties in laryngeal squamous cell carcinoma via E2F1-mediated upregulation of HMGB3. Exp. Mol. Med. 2021, 53, 1218–1228.
- Wang, X.; Meng, R.; Hu, Q.-M. LINC00319-Mediated miR-3127 Repression Enhances Bladder Cancer Progression Through Upregulation of RAP2A. Front. Genet. 2020, 11, 180.
- Luo, X.; Abudureyimu, M.; Yang, G.; Yan, Z.; Fu, X.; Lu, P.; Zhang, D.; Zhang, S.; Ding, Z. LINC00355 triggers malignant progression of hepatocellular carcinoma via the sponge effect on miR-217-5p with the involvement of the Wnt/β-catenin signaling. J. BUON 2021, 26, 1964–1969.
- Qi, Z.-Y.; Wang, L.-L.; Qu, X.-L. lncRNA LINC00355 Acts as a Novel Biomarker and Promotes Glioma Biological Activities via the Regulation of miR-1225/FNDC3B. Dis. Markers 2021, 2021, 1683129.
- Sun, X.; Wang, G.; Ding, P.; Li, S. LINC00355 promoted the progression of lung squamous cell carcinoma through regulating the miR-466/LYAR axis. Braz. J. Med. Biol. Res. 2020, 53, e9317.
- Li, W.-J.; Li, G.; Liu, Z.-W.; Chen, Z.-Y.; Pu, R. LncRNA LINC00355 promotes EMT and metastasis of bladder cancer cells through the miR-424-5p/HMGA2 axis. Neoplasma 2021, 68, 1225–1235.
- Zhu, H.; Liu, Q.; Yang, X.; Ding, C.; Wang, Q.; Xiong, Y. LncRNA LINC00649 recruits TAF15 and enhances MAPK6 expression to promote the development of lung squamous cell carcinoma via activating MAPK signaling pathway. Cancer Gene Ther. 2022, 29, 1285–1295.
- Zhang, J.; Du, C.; Zhang, L.; Wang, Y.; Zhang, Y.; Li, J. LncRNA LINC00649 promotes the growth and metastasis of triple-negative breast cancer by maintaining the stability of HIF-1α through the NF90/NF45 complex. Cell Cycle 2022, 21, 1034–1047.
- Wang, H.; Di, X.; Bi, Y.; Sun, S.; Wang, T. Long non-coding RNA LINC00649 regulates YES-associated protein 1 (YAP1)/Hippo pathway to accelerate gastric cancer (GC) progression via sequestering miR-16-5p. Bioengineered 2021, 12, 1791–1802.
- Feng, L.; Yang, J.; Zhang, W.; Wang, X.; Li, L.; Peng, M.; Luo, P. Prognostic significance and identification of basement membrane-associated lncRNA in bladder cancer. Front. Oncol. 2022, 12, 994703.
- Li, H.; Gao, J.; Liu, L.; Zhang, S. LINC00958: A promising long non-coding RNA related to cancer. Biomed. Pharmacother. 2022, 151, 113087.
