You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 3 by Jessie Wu and Version 4 by Jessie Wu.
Filamentous fungi are an important source of natural products. The mold Penicillium roqueforti, which is well-known for being responsible for the characteristic texture, blue-green spots, and aroma of the so-called blue-veined cheeses (French Bleu, Roquefort, Gorgonzola, Stilton, Cabrales, and Valdeón, among others), is able to synthesize different secondary metabolites, including andrastins and mycophenolic acid, as well as several mycotoxins, such as Roquefortines C and D, PR-toxin and eremofortins, Isofumigaclavines A and B, festuclavine, and Annullatins D and F.
- Penicillium roqueforti
- blue cheese
- secondary metabolism
1. Brief Overview of Taxonomic and Biotechnological Aspects of Penicillium roqueforti
Penicillium roqueforti (P. roqueforti)is a saprophytic filamentous fungus (mold) whose colonies show a color from light to dark greenish gray and can include gray, yellowish, and olive-green shades. It also has a texture that can vary from velvety to fasciculate. Conidiophores constitute a velutinous felt with phialides, which produce spherical, smooth, and dark green conidia (3 to 4.5 μm diameter) included in terminal penicilli, which are typically terverticillate (quaterverticillate and more rarely biverticillate can also be observed) [1].
The original taxonomic description of the species P. roqueforti was performed by Thom in 1906 [2], using a strain isolated from a Roquefort cheese purchased in a market in the United States. Despite this early taxonomic description, the taxonomy of this species has been complex. In the past, the denomination P. roqueforti included a group of heterogeneous fungi (the “P. roqueforti group”) with very similar phenotypic characteristics, which are hard to distinguish by the traditional morphological and physiological methods [3]. In addition, through the years, numerous strains of P. roqueforti independently isolated by different researchers were designated with different names, making more complex the establishment of accurate taxonomic denominations for new isolates. With the advent of molecular techniques, the taxonomy of the “P. roqueforti group” began to be clarified [3]. In 2004, Frisvad and Samson accomplished the full taxonomy of this species synonymizing many of the different names given to P. roqueforti over the years and designating P. roqueforti IMI 024313 as the neotype for the species [4].
Currently, P. roqueforti is classified within the subgenus Penicillium, section Roquefortorum, and series Roquefortorum along the closely related species P. carneum, P. mediterraneum, P. paneum, and P. psychrosexuale [5]. As all the members of this series, P. roqueforti is characterized by having large globose conidia and rough-walled conidiophore stipes, the ability to grow at elevated carbon dioxide levels, and the ability to produce the mycotoxin roquefortine C, but it can be clearly distinguished from the other members of the series by phylogenetic analyses using concatenated markers, its ability to produce specific secondary metabolites, and its capability of performing heterothallic sexual reproduction [5][6].
P. roqueforti has been used for centuries as a maturation agent in blue cheese, which includes several varieties such as French Bleu and Roquefort, Italian Gorgonzola, English Stilton, Spanish Cabrales, Picón Bejes-Tresviso and Valdeón, and many others from Denmark and the United States [1][7]. Blue cheese receives this name due to the blue-veined appearance that results from the proliferation of the melanized fungal conidia (asexual spores) within aerated cavities in the cheese [8]. P. roqueforti spores spontaneously contaminating milk were the origin of blue cheese. However, since the end of the 18th century, conidia are inoculated during the production process [9][10], which is slightly different according to the variety. This process mainly comprises the inoculation of the ripening mold P. roqueforti in liquid suspensions, which are added either to milk batches containing high levels of fat obtained from sheep or cow, depending on the variety, or the curd. Fermentation of this type of cheese is carried out by mesophilic lactic acid bacteria: Streptococcus lactis, Streptococcus lactis subsp. diacetylactis and Leuconostoc spp. [7]. In addition, P. roqueforti acts as a secondary starter providing cheese with a characteristic intense and spicy flavour because of the proteolytic and lipolytic activities, which, during ripening, generate volatile and non-volatile compounds responsible for the mouldy aromas [11][12]. During the maturation process, other microorganisms, such as Brevibacterium linens, also proliferate and add specific aroma to many blue cheeses [13].
In addition to the main use of this microorganism in the production of blue cheese, P. roqueforti has also been considered for other biotechnological purposes, such as the production of different metabolites, including the immunosuppressant agent mycophenolic acid [14], lipase extracts on solid state fermentation using cocoa shells as a substrate [15], or cellulolytic enzyme extracts upon cultivation on residue of yellow mombin fruit [16].
2. Secondary Metabolites Produced by Penicillium roqueforti
For many years, P. roqueforti has been known to produce several secondary metabolites with biological properties, which encouraged the chemical study of this fungal species. As a result, numerous secondary metabolites were purified for the first time from P. roqueforti cultures, and their structures were clarified. In addition, other compounds already known to be produced by other fungi are also produced by P. roqueforti. A brief overview of these secondary metabolites (Figure 1) is described below.
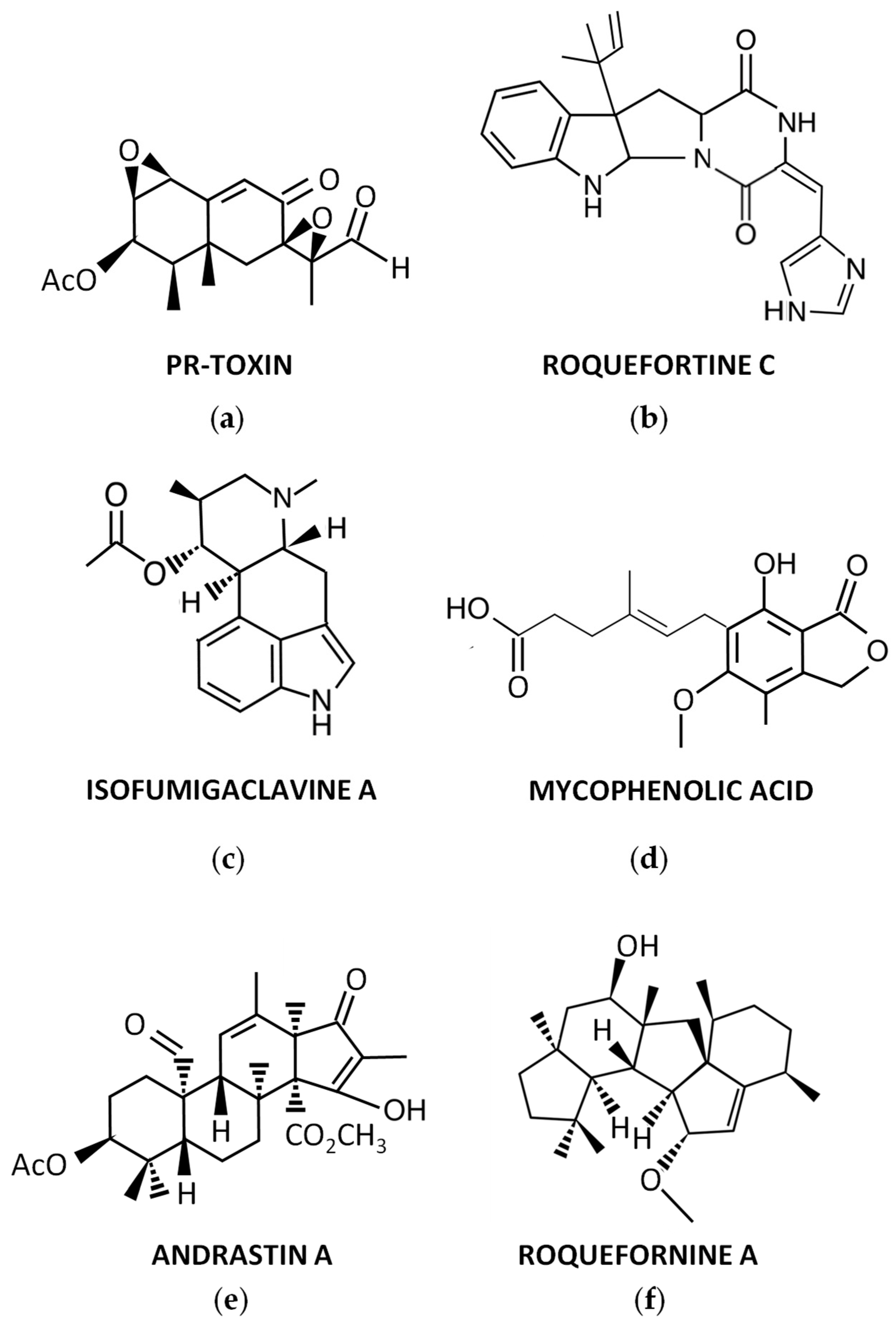
Figure 1. Representative secondary metabolites produced by P. roqueforti: (a) PR-toxin; (b) Roquefortine C; (c) Isofumigaclavine A; (d) Mycophenolic acid; (e) Andrastin A; (f) Roquefornine A.
2.1. PR-Toxin and Related Compounds
During the 70s, a potent toxin from P. roqueforti, named PR-toxin (Figure 1a), was discovered by Wei et al. [17]. The compound was purified from stationary cultures of the fungus on liquid YES medium (2% yeast extract, 15% sucrose), and its toxicity to rats was demonstrated [17]. In this original description, the chemical structure of PR-toxin was only partially elucidated. The full elucidation of its structure was achieved in a subsequent work [18]. From a chemical point of view, PR-toxin belongs to the eremophilane terpenoid class, it is a bicyclic sesquiterpene with the presence of two stable epoxide rings, and it has several functional groups [19].
PR-toxin is unstable and can be readily converted into other compounds. This property led to the identification of some PR-toxin related metabolites. Chang et al. [20] found that two compounds appeared in the culture medium of P. roqueforti, while PR toxin decreased. These compounds were purified, and their structures were elucidated, revealing that they were PR-imine and PR-amide (also known as Eremofortin E) [20]. Later, the same group purified and identified a third degradation product, which was named PR-acid [21].
As was mentioned before, PR-toxin belongs to the eremophilane terpenoid class. Accordingly, eremophilane compounds related to PR-toxin have also been purified and characterized. In this way, in three consecutive papers, Moreau and co-workers reported five eremofortins (A–E) related to PR-toxin, which were isolated from filtrates of P. roqueforti [22][23][24].
2.2. Roquefortine C and Related Compounds
Roquefortines are a family of prenylated diketopiperazine indole alkaloids, whose core structure is formed by the condensation of L-tryptophan and L-histidine [25]. Roquefortines are produced by several fungi from the genus Penicillium [26]. The main member of this family is Roquefortine C (Figure 1b), which is also one of the major secondary metabolites produced by P. roqueforti [25]. Roquefortine C was originally isolated from cultures of P. roqueforti in 1975 [27], although its full structural elucidation was achieved a couple of years after [28][29].
In addition to roquefortine C, the other member of the family that has been isolated from cultures of Penicillium roqueforti is Roquefortine D [30]. At this point, it should be mentioned that there are other members of the roquefortine family as well as related compounds (meleagrin, glandicolins), but they are not produced by P. roqueforti [25]. This point will be revisited below.
2.3. Other Secondary Metabolites Produced by Penicillium roqueforti
PR-toxin and Roquefortine C are the main toxic secondary metabolites produced by P. roqueforti, so they were treated separately before. However, this fungus can produce other minor mycotoxins as well as non-toxic compounds, which are briefly described in this section.
In early studies on purification of Roquefortine C, some ergot alkaloids were co-purified [27]. They included festuclavine and two other compounds that were originally named Roquefortine A and Roquefortine B, which correspond to the ergot alkaloids Isofumigaclavine A (Figure 1c) and Isofumigaclavine B, respectively [27]. In addition, another ergot alkaloid, agroclavine, was detected by mass spectrometry in P. roqueforti [31].
Another interesting compound produced by P. roqueforti is mycophenolic acid (Figure 1d). This compound is a meroterpenoid with a phthalide moiety that exhibits several biological activities, being widely used as immunosuppressant for the prevention of organ transplant rejection [32]. Mycophenolic acid is commonly detected in many P. roqueforti strains [31][33].
Andrastins A–D are a family of meroterpenoid compounds derived from dimethyl orsellinic acid, which are interesting candidates as anticancer drugs [25]. Andrastins, especially the main member of the family, Andrastin A (Figure 1e), are usually detected in P. roqueforti [31][33][34].
In recent years, a family of sesterterpenoid compounds were isolated from P. roqueforti. The first compounds isolated from this family were the pentacyclic sesterterpenes Peniroquesines A–C [35]. Later, Roquefornine A (Figure 1f), another sesterterpene with an unprecedented pentacyclic system and cytotoxic activity, was described by Wang et al. [36]. Finally, seven other compounds related to Peniroquesines A–C were recently described, some of them showing interesting cytotoxic and anti-inflammatory activities [37].
3. Control and Regulation of the Biosynthesis of Secondary Metabolites in Penicillium roqueforti
The fungal secondary metabolism is controlled by a wide array of regulators, which lead directly or indirectly to the activation or repression of a given biosynthetic gene cluster. Depending on the level and extension of the effect, regulators of fungal secondary metabolism can be classified in two main groups: cluster-specific regulators and global regulators [38]. A cluster-specific regulator (also named specific transcription factor or narrow domain regulator) refers to a regulatory protein with a high similarity to a transcription factor that is encoded by a gene that is part of a specific biosynthetic gene cluster. More importantly, this transcription factor specifically regulates the expression of the other genes of the biosynthetic gene cluster [38]. On the contrary, global regulators (also named wide domain regulators) are encoded by genes located outside of a biosynthetic gene cluster and are pleiotropic regulators in fungi. Although most of these regulators are transcription factors, this category also includes signal transducer proteins and epigenetic regulators [38]. In the following sections, rwesearchers will summarize the current knowledge about the regulation of biosynthetic gene clusters in P. roqueforti by cluster-specific and global regulators.3.1. Cluster-Specific Regulators in Biosynthetic Gene Clusters Functionally Characterized in Penicillium roqueforti
As mentioned before, the biosynthetic gene clusters for the biosynthesis of PR-toxin, Andrastin A, Roquefortine C, mycophenolic acid, annullatins, and Isofumigaclavine A have been functionally characterized in P. roqueforti [39][40][41][42][43][44][45][46]. Among them, only the gene clusters for the biosynthesis of annullatins and PR-toxin contain candidate genes for a specific transcription factor [42][46]. In the case of the biosynthetic gene cluster of Annullatin D, the functionality of the gene encoding the specific transcription factor (named anuK) was partially demonstrated [46]. As mentioned, the Annullatin D biosynthetic gene cluster seems to be silent. Therefore, it was heterologously expressed in A. nidulans for its functional characterization. For this purpose, the biosynthetic gene cluster was previously cloned into plasmid pJN017, which contains the constitutive promoter of the gpdA gene. Therefore, the authors took advantage of the fact that the first gene of the biosynthetic gene cluster corresponds to anuK and cloned it without the natural promoter of anuK in such a way that this gene was under the control of the gpdA promoter [46]. As a result, the production of Annullatin D, Annullatin F, and other related metabolites was achieved. At this point, it should be noted that beyond this result, there is no further experimental evidence confirming the role of AnuK as the transcriptional activator of the annullatin biosynthetic gene cluster in P. roqueforti. However, bioinformatic analysis supports such a role. AnuK has high similarity to FogI, a putative transcription factor found in the biosynthetic gene cluster for the biosynthesis of flavoglaucin in A. ruber [47]. In addition, AnuK contains a Zn(II)2Cys6 domain. This kind of domain is found almost exclusively in fungi and has been linked to several functions, including the regulation of the secondary metabolism [48][49]. Concerning the biosynthetic gene cluster of PR-toxin, its functional characterization in P. roqueforti was reported in two consecutive papers [41][42], where seven genes were silenced by RNAi-mediated gene-silencing technology. Unfortunately, the gene encoding the putative transcription factor of the biosynthetic gene cluster, named ORF10 (Figure 4) [42], was not analyzed. Similar to AnuK described before, ORF10 encodes a protein that contains a Zn(II)2Cys6 domain. ORF10 is highly similar to orthologous genes found in the biosynthetic gene clusters for PR-toxin in P. camemberti and P. chrysogenum [42]. Thus, the high degree of conservation of ORF10 in biosynthetic gene clusters of different fungi suggests that this gene could be functional. In the future it will be interesting to determine the role of ORF10 in the regulation of the biosynthetic gene cluster for the biosynthesis of PR-toxin in P. roqueforti.3.2. Global Regulators of Biosynthetic Gene Clusters in Penicillium roqueforti
Almost all the current knowledge about the regulation of biosynthetic gene clusters in P. roqueforti corresponds to global regulators. However, it should be kept in mind that in comparison with model fungi (Aspergillus spp., P. chrysogenum, etc.), this knowledge is still scarce. Indeed, as far as we know, the effect of only three global regulators on the secondary metabolism have been studied in P. roqueforti. They are detailed below.3.2.1. The pga1 Gene Encoding for an α-Subunit of a Heterotrimeric G Protein
The first global regulator studied in P. roqueforti was a heterologous gene named pga1, which encodes an α-subunit of a heterotrimeric G protein. Heterotrimeric G proteins, particularly α-subunits, are important in fungi and have been involved mainly in the regulation of growth, asexual reproduction, and secondary metabolism [50]. García–Rico et al. [51] isolated and characterized the pga1 gene from P. chrysogenum, and they decided to perform the heterologous expression of a mutant version of this gene in P. roqueforti [52]. For this purpose, they used a plasmid containing a mutant version of pga1 (named pga1G42R), which encodes a protein where a glycine is replaced by arginine at Position 42, producing a “constitutively active” Pga1 protein that is always signaling [53]. The introduction of this “constitutive” allele in P. roqueforti produced several phenotypic effects. In the case of secondary metabolism, García–Rico et al. [52] measured the production of Roquefortine C during 30 days in a strain of P. roqueforti containing the pga1G42R allele and observed drastic changes in the production of this secondary metabolite as compared with the wild-type strain. Namely, the strain containing pga1G42R increased production of the mycotoxin, reaching 0.7 μg/mg of dry mycelium at day 18, whereas the wild-type strain produced 0.4 μg/mg of dry mycelium at the same day. Interestingly, the levels of Roquefortine C were higher in the pga1G42R strain throughout a culture period ranging between 10–30 days [52]. These results suggested that pga1 has a positive effect on the production of Roquefortine C in P. roqueforti [52]. As mentioned above, these experiments were performed using a heterologous gene from P. chrysogenum. Therefore, the role of the native pga1 gene on the secondary metabolism of P. roqueforti remains to be tested.3.2.2. The sfk1 Gene Encoding for Suppressor of Four-Kinase 1 Protein
Another global regulator influencing the biosynthesis of secondary metabolites in P. roqueforti is Sfk1 (Suppressor of four-kinase 1), a transmembrane protein located on the plasma membrane that was originally described in Saccharomyces cerevisiae [54]. In this yeast, Sfk1 physically interacts with Stt4, an important phosphatidylinositol 4-kinase involved in the phosphoinositide second messenger’s pathway [55]. Stt4 must be localized to the plasma membrane to fulfill its role. Hence, it is thought that its interaction with Sfk1 would allow its proper subcellular localization [56]. In addition, Sfk1 is essential for the retention of ergosterol in the plasma membrane of the yeast [57]. The mammalian homologue of Sfk1 was also studied [58][59]. The role of this homologue is essentially similar to that described in yeast, although it does not have a direct role in mammals in the localization of the phosphatidylinositol 4-kinase to the plasma membrane [58]. In addition, it has been suggested that in mammals, Sfk1 would also be a negative regulator of transbilayer movement of phospholipids in plasma membrane [59]. The interest in studying Sfk1 in P. roqueforti emerged from a previous work by Gil-Durán and co-workers [60]. These authors performed suppression subtractive hybridization (SSH) experiments, looking for downstream genes regulated by Pga1. Among the sequences obtained from these experiments, the full cDNA of sfk1 gene was found. Considering this antecedent, Torrent and co-workers [61] decided to analyze the role of this gene in P. roqueforti. For this purpose, they performed the RNA-mediated gene-silencing of sfk1 and measured several biological properties in the transformants. In the case of secondary metabolites, they observed that the knock-down of sfk1 produced a drastic decrease in the production of Roquefortine C, Andrastin A, and mycophenolic acid in P. roqueforti [61]. These results suggest that sfk1 is a positive regulator of the production of these three secondary metabolites in P. roqueforti.3.2.3. The pcz1 Gene Encoding a Protein with a Zn(II)2Cys6 Domain
The last global regulator of secondary metabolism studied in P. roqueforti is that encoded by pcz1 (Penicillium C6 zinc-finger protein 1), a gene whose role in fungi was unknown until recent time. This gene encodes a protein containing a Zn(II)2Cys6 domain [60]. As in the case of sfk1, pcz1 was also obtained from SSH experiments performed on P. roqueforti [60]. Since there are orthologues of this gene in all ascomycetes analyzed so far [60], it seemed interesting to determine its role on the secondary metabolism of P. roqueforti. For this purpose, two kinds of P. roqueforti strains were obtained: strains overexpressing pcz1 [62] and strains where pcz1 was knocked-down by RNA-silencing technology [60]. Using both types of strains, the production of Roquefortine C, Andrastin A, and mycophenolic acid was measured in comparison to the wild-type strain [62]. In the case of mycophenolic acid, a clear effect was found. Strains overexpressing pcz1 showed higher titers of mycophenolic acid than the wild-type strain, which correlated with higher levels of the expression of key genes from the mycophenolic acid biosynthetic gene cluster [62]. On the contrary, strains where pcz1 was knocked down produced lower levels of the mycophenolic acid as compared to wild-type fungus in concomitance with lower levels of the expression of genes from its biosynthetic gene cluster [62]. These results indicate that pcz1 exerts a positive control of the production of mycophenolic acid and suggest that this effect is mediated by modifying the transcriptional status of the biosynthetic gene cluster of mycophenolic acid. Unlike mycophenolic acid, important reductions in the production of Roquefortine C and Andrastin A were observed in both types of strains [62]. These results were confirmed by measuring the level of transcription of key genes from Roquefortine C and Andrastin A biosynthetic gene clusters [62]. Thus, the effect of pcz1 on these metabolites was unexpected and difficult to interpret. Considering these results, an indirect effect of pcz1 on the production of Roquefortine C and Andrastin A was suggested, and some hypotheses were proposed. The first one was related to competition for the use of sharing substrates. These three compounds require acetyl CoA directly or indirectly for their biosynthesis. Therefore, the overexpression of the mycophenolic acid pathway (produced in turn by the overexpression of pcz1) would use more efficiently acetyl-CoA, in detriment of the production of Andrastin A and Roquefortine C. This would produce a “negative loop” in the expression of Andrastin A and Roquefortine C biosynthetic gene clusters, thereby resulting in a low production of these compounds. The second hypothesis was the alteration of the normal balance of unknown regulators of Roquefortine C and Andrastin A because of the overexpression of pcz1 [62]. A similar phenomenon was described before in Aspergillus. In this case, it was observed that the elimination of the regulator mtfA decreased the production of two related mycotoxins: aflatoxin and sterigmatocystin [63][64]. However, the overexpression of mtfA also decreased the production of these mycotoxins because mtfA downregulates the normal expression of the transcriptional factor aflR, which is a positive activator of the biosynthesis of aflatoxin and sterigmatocystin in Aspergillus [63][64]. Finally, the last hypothesis suggested the existence of a regulatory mechanism not fully understood, known as regulatory cross-talk [62]. Regulatory cross-talk refers to an inter-regulation amongst different and non-related biosynthetic gene clusters in fungi [65]. Regulatory cross-talk was originally described in A. nidulans, where the overexpression of a gene encoding a NRPS of the so-called inp biosynthetic gene cluster resulted in the unexpected activation of the non-related biosynthetic gene cluster for the biosynthesis of asperfuranone [66]. There are several other examples of regulatory cross-talk in different fungi [65]. In P. roqueforti, regulatory cross-talk has been described in the case of biosynthetic gene clusters encoding PR-toxin and mycophenolic acid [41]. More precisely, during the experiments of the functional characterization of the PR-toxin biosynthetic gene cluster [41], it was noticed that the down regulation of several genes of this biosynthetic gene cluster by RNAi-mediated silencing technology largely increased the production of mycophenolic acid. It is important to highlight that PR-toxin and mycophenolic acid are unrelated secondary metabolites, so this result can hardly be due to an imbalance of shared substrates. PR-toxin is a bicyclic sesquiterpene that belongs to eremophilane. Its core structures is aristolochene, a sesquiterpene produced from farnesyl diphosphate [19], whereas mycophenolic acid is a meroterpenoid whose core structure, 5-methylorsellinic acid (5-MOA), is formed by a polyketide synthase. 5-MOA is subjected to several further modifications (including prenylation) to yield mycophenolic acid [67]. Thus, both compounds are synthesized by entirely different biosynthetic pathways in P. roqueforti [42][43]. Considering the previous observation of regulatory cross-talk in P. roqueforti, Rojas–Aedo and co-workers [62] suggested that the unexpected result in the regulation of mycophenolic acid, Andrastin A and Roquefortine C by pcz1 in this fungus, could be due to the existence of regulatory cross-talk between the biosynthetic gene clusters of these metabolites. Hence, when pcz1 is subjected to genetic manipulation, a complex network of regulatory cross-talk between these biosynthetic gene clusters is triggered. Depending on the case, the overproduction or reduction in the levels of some compounds is observed. However, this remains as a hypothesis that requires further experimental support.3.3. Concluding Remarks on Regulation of Secondary Metabolism in Penicillium roqueforti
In comparison with other fungi, our current knowledge about the regulation of the biosynthesis of secondary metabolites in P. roqueforti is poor, which should encourage fungal biologists to pay more attention to this interesting topic. As mentioned above, to date, barely one specific regulator of a biosynthetic gene cluster has been indirectly characterized in P. roqueforti [46], and some important global regulators, such as CreA, LaeA, PacC, or AreA have not yet been analyzed in this fungus. This is unlike in other fungi, where their role in the control of secondary metabolism has been largely demonstrated. For example, CreA exerts carbon catabolic repression on penicillin biosynthesis and the expression of the pcbAB (the gene encoding the first enzyme of the penicillin pathway) in P. chrysogenum [68], while PacC exerts pH-dependent control on the production of patulin and the expression of its BGC in P. expansum [69]. In the case of LaeA, its influence on the secondary metabolism of several Penicillium species has been documented [38]. Concerning the regulator of the nitrogen metabolism AreA, it has been involved in the control of the production of secondary metabolites in P. chrysogenum and P. griseofulvum [38]. Interestingly, the areA gene homologue has been studied in P. roqueforti [70], but its role on secondary metabolism has not been addressed in this fungus.References
- Coton, E.; Coton, M.; Hymery, N.; Mounier, J.; Jany, J.-L. Penicillium roqueforti: An overview of its genetics, physiology, metabolism and biotechnological applications. Fungal Biol. Rev. 2020, 34, 59–73.
- Thom, C. Fungi in cheese ripening: Camembert and Roquefort. USDA Bureau Anim. Industry Bull. 1906, 82, 1–39.
- Boysen, M.; Skouboe, P.; Frisvad, J.; Rossen, L. Reclassification of the Penicillium roqueforti group into three species on the basis of molecular genetic and biochemical profiles. Microbiology 1996, 142, 541–549.
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium—A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–73.
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169.
- Ropars, J.; López-Villavicencio, M.; Dupont, J.; Snirc, A.; Gillot, G.; Coton, M.; Jany, J.L.; Coton, E.; Giraud, T. Induction of sexual reproduction and genetic diversity in the cheese fungus Penicillium roqueforti. Evol. Appl. 2014, 7, 433–441.
- Albillos, S.M.; García-Estrada, C.; Martín, J.-F. Spanish blue cheeses: Functional metabolites. In Cheese: Types, Nutrition and Consumption; Foster, R.D., Ed.; Nova Science Publishers, Inc.: New York, NY, USA, 2011; pp. 89–105.
- Moreau, C. Le Penicillium roqueforti, morphologie, physiologie, interet en industrie fromagere, mycotoxines. Lait 1980, 60, 254–271.
- Labbe, M.; Serres, J.-P. Chroniques du Roquefort: De la préhistoire à l′aube de l′ère industrielle; Graphi Imprimeur: La Primaube, France, 2004.
- Labbe, M.; Serres, J.-P. Chroniques du Roquefort: Des hommes, des entreprises, des marques, période modern; Graphi Imprimeur: La Primaube, France, 2009.
- McSweeney, P.L.H.; Sousa, M.J. Biochemical pathways for the production of flavour compounds in cheeses during ripening: A review. Lait 2000, 80, 293–324.
- Collins, Y.F.; McSweeney, P.L.H.; Wilkinson, M.G. Lipolysis and free fatty acid catabolism in cheese: A review of current knowledge. Int. Dairy J. 2003, 13, 841–866.
- Deetae, P.; Bonnarme, P.; Spinnler, H.E.; Helinck, S. Production of volatile aroma compounds by bacterial strains isolated from different surface-ripened French cheeses. Appl. Microbiol. Biotechnol. 2007, 76, 1161–1171.
- Ismaiel, A.A.; Ahmed, A.S.; El-Sayed, E.S.R. Optimization of submerged fermentation conditions for immunosuppressant mycophenolic acid production by Penicillium roqueforti isolated from blue-molded cheeses: Enhanced production by ultraviolet and gamma irradiation. World J. Microbiol. Biotechnol. 2014, 30, 2625–2638.
- Silva, T.P.; Souza, L.O.; Reis, N.S.; Assis, S.A.; Ferreira, M.L.O.; Oliveira, J.R.; Aguiar-Oliveira, E.; Franco, M. Cultivation of Penicillium roqueforti in cocoa shell to produce and characterize its lipase extract. Rev. Mex. Ing. Quim. 2017, 16, 745–756.
- de Almeida Antunes Ferraz, J.L.; Souza, L.O.; Soares, G.A.; Coutinho, J.P.; de Oliveira, J.R.; Aguiar-Oliveira, E.; Franco, M. Enzymatic saccharification of lignocellulosic residues using cellulolytic enzyme extract produced by Penicillium roqueforti ATCC 10110 cultivated on residue of yellow mombin fruit. Bioresour. Technol. 2018, 248, 214–220.
- Wei, R.D.; Still, P.E.; Smalley, E.B.; Schnoes, H.K.; Strong, F.M. Isolation and partial characterization of a mycotoxin from Penicillium roqueforti. Appl. Microbiol. 1973, 25, 111–114.
- Wei, R.D.; Schnoes, H.K.; Hart, P.A.; Strong, F.M. The structure of PR toxin, a mycotoxin from Penicillium roqueforti. Tetrahedron 1975, 31, 109–114.
- Dubey, M.K.; Aamir, M.; Kaushik, M.S.; Khare, S.; Meena, M.; Singh, S.; Upadhyay, R.S. PR toxin—Biosynthesis, genetic regulation, toxicological potential, prevention and control measures: Overview and challenges. Front. Pharmacol. 2018, 9, 288.
- Chang, S.C.; Lu, K.L.; Yeh, S.F. Secondary metabolites resulting from degradation of PR toxin by Penicillium roqueforti. Appl. Environ. Microbiol. 1993, 59, 981–986.
- Chang, S.C.; Yeh, S.F.; Li, S.Y.; Lei, W.Y.; Chen, M.Y. A novel secondary metabolite relative to the degradation of PR toxin by Penicillium roqueforti. Curr. Microbiol. 1996, 32, 141–146.
- Moreau, S.; Gaudemer, A.; Lablache-Combier, A.; Biguet, J. Metabolites de Penicillium roqueforti: PR toxine et metabolites associes. Tetrahedron Lett. 1976, 11, 833–834.
- Moreau, S.; Cacan, M.; Eremofortin, C. A new metabolite obtained from Penicillium roqueforti cultures and from biotransformation of PR toxin. J. Org. Chem. 1977, 42, 2632–2634.
- Moreau, S.; Biguet, J.; Lablache-Combier, A.; Baert, M.; Delfosse, C. Structures et stereochimie des sesquiterpenes de Penicillium roqueforti pr toxine et eremofortines a, b, c, d, e. Tetrahedron. 1980, 36, 2989–2997.
- García-Estrada, C.; Martín, J.F. Biosynthetic gene clusters for relevant secondary metabolites produced by Penicillium roqueforti in blue cheeses. Appl. Microbiol. Biotechnol. 2016, 100, 8303–8313.
- Frisvad, J.C.; Smedsgaard, J.; Larsen, T.O.; Samson, R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud. Mycol. 2004, 49, 201–241.
- Ohmomo, S.; Sato, T.; Utagawa, T.; Abe, M. Isolation of festuclavine and three new indole alkaloids, roquefortine A, B and C from the cultures of Penicillium roqueforti. Agr. Biol. Chem. 1975, 39, 1333–1334.
- Ohmomo, S.; Utagawa, T.; Abe, M. Identification of roquefortine C produced by Penicillium roqueforti. Agr. Biol. Chem. 1977, 41, 2097–2098.
- Scott, P.M.; Merrien, M.A.; Polonsky, J. Roquefortine and isofumigaclavine A, metabolites from Penicillium roqueforti. Experientia 1976, 32, 140–142.
- Ohmomo, S.; Oguma, K.; Ohashi, T.; Abe, M. Isolation of a new indole alkaloid, roquefortine D, from the cultures of Penicillium roqueforti. Agr. Biol. Chem. 1978, 42, 2387–2389.
- Nielsen, K.F.; Sumarah, M.W.; Frisvad, J.C.; Miller, J.D. Production of metabolites from the Penicillium roqueforti complex. J. Agric. Food Chem. 2006, 54, 3756–3763.
- Bentley, R. Mycophenolic acid: A one hundred year odyssey from antibiotic to immunosuppressant. Chem. Rev. 2000, 100, 3801–3826.
- O′Brien, M.; Nielsen, K.F.; O′Kiely, P.; Forristal, P.D.; Fuller, H.T.; Frisvad, J.C. Mycotoxins and other secondary metabolites produced in vitro by Penicillium paneum Frisvad and Penicillium roqueforti Thom isolated from baled grass silage in Ireland. J. Agric. Food Chem. 2006, 54, 9268–9276.
- Nielsen, K.F.; Dalsgaard, P.W.; Smedsgaard, J.; Larsen, T.O. Andrastins A-D, Penicillium roqueforti metabolites consistently produced in blue-mold-ripened cheese. J. Agric. Food Chem. 2005, 53, 2908–2913.
- Wang, J.P.; Yu, J.; Shu, Y.; Shi, Y.X.; Luo, P.; Cai, L.; Ding, Z.T. Peniroquesines A-C: Sesterterpenoids possessing a 5-6-5-6-5-fused pentacyclic ring system from Penicillium roqueforti YJ-14. Org. Lett. 2018, 20, 5853–5856.
- Wang, J.P.; Shu, Y.; Hu, J.T.; Liu, R.; Cai, X.Y.; Sun, C.T.; Gan, D.; Zhou, D.J.; Mei, R.F.; Ding, H.; et al. Roquefornine A, a sesterterpenoid with a 5/6/5/5/6-fused ring system from the fungus Penicillium roqueforti YJ-14. Org. Chem. Front. 2020, 7, 1463–1468.
- Wang, J.P.; Shu, Y.; Liu, R.; Gan, J.L.; Deng, S.P.; Cai, X.Y.; Hu, J.T.; Cai, L.; Ding, Z.T. Bioactive sesterterpenoids from the fungus Penicillium roqueforti YJ-14. Phytochemistry 2021, 187, 112762.
- El Hajj Assaf, C.; Zetina-Serrano, C.; Tahtah, N.; Khoury, A.E.; Atoui, A.; Oswald, I.P.; Puel, O.; Lorber, S. Regulation of secondary metabolism in the Penicillium genus. Int. J. Mol. Sci. 2020, 21, 9462.
- Kosalková, K.; Domínguez-Santos, R.; Coton, M.; Coton, E.; García-Estrada, C.; Liras, P.; Martín, J.F. A natural short pathway synthesizes roquefortine C but not meleagrin in three different Penicillium roqueforti strains. Appl. Microbiol. Biotechnol. 2015, 99, 7601–7612.
- Fernández-Bodega, Á.; Álvarez-Álvarez, R.; Liras, P.; Martín, J.F. Silencing of a second dimethylallyltryptophan synthase of Penicillium roqueforti reveals a novel clavine alkaloid gene cluster. Appl. Microbiol. Biotechnol. 2017, 101, 6111–6121.
- Hidalgo, P.I.; Ullán, R.V.; Albillos, S.M.; Montero, O.; Fernández-Bodega, M.Á.; García-Estrada, C.; Fernández-Aguado, M.; Martín, J.F. Molecular characterization of the PR-toxin gene cluster in Penicillium roqueforti and Penicillium chrysogenum: Cross talk of secondary metabolite pathways. Fungal Genet. Biol. 2014, 62, 11–24.
- Hidalgo, P.I.; Poirier, E.; Ullán, R.V.; Piqueras, J.; Meslet-Cladière, L.; Coton, E.; Coton, M. Penicillium roqueforti PR toxin gene cluster characterization. Appl. Microbiol. Biotechnol. 2017, 101, 2043–2056.
- Del-Cid, A.; Gil-Durán, C.; Vaca, I.; Rojas-Aedo, J.F.; García-Rico, R.O.; Levicán, G.; Chávez, R. Identification and functional analysis of the mycophenolic acid gene cluster of Penicillium roqueforti. PLoS ONE 2016, 11, e0147047.
- Gillot, G.; Jany, J.L.; Dominguez-Santos, R.; Poirier, E.; Debaets, S.; Hidalgo, P.I.; Ullán, R.V.; Coton, E.; Coton, M. Genetic basis for mycophenolic acid production and strain-dependent production variability in Penicillium roqueforti. Food Microbiol. 2017, 62, 239–250.
- Rojas-Aedo, J.F.; Gil-Durán, C.; Del-Cid, A.; Valdés, N.; Álamos, P.; Vaca, I.; García-Rico, R.O.; Levicán, G.; Tello, M.; Chávez, R. The biosynthetic gene cluster for andrastin A in Penicillium roqueforti. Front. Microbiol. 2017, 8, 813.
- Xiang, P.; Kemmerich, B.; Yang, L.; Li, S.M. Biosynthesis of annullatin D in Penicillium roqueforti implies oxidative lactonization between two hydroxyl groups catalyzed by a BBE-like enzyme. Org. Lett. 2022, 24, 6072–6077.
- Nies, J.; Ran, H.; Wohlgemuth, V.; Yin, W.B.; Li, S.M. Biosynthesis of the prenylated salicylaldehyde flavoglaucin requires temporary reduction to salicyl alcohol for decoration before reoxidation to the final product. Org. Lett. 2020, 22, 2256–2260.
- Chang, P.K.; Ehrlich, K.C. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2013, 97, 4289–4300.
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.-F. Transcription factors controlling primary and secondary metabolism in filamentous fungi: The β-lactam paradigm. Fermentation 2018, 4, 47.
- Moon, H.; Han, K.H.; Yu, J.H. Upstream regulation of development and secondary metabolism in Aspergillus species. Cells 2022, 12, 2.
- García-Rico, R.O.; Martín, J.F.; Fierro, F. The pga1 gene of Penicillium chrysogenum NRRL 1951 encodes a heterotrimeric G protein alpha subunit that controls growth and development. Res. Microbiol. 2007, 158, 437–446.
- García-Rico, R.O.; Chávez, R.; Fierro, F.; Martín, J.F. Effect of a heterotrimeric G protein alpha subunit on conidia germination, stress response, and roquefortine C production in Penicillium roqueforti. Int. Microbiol. 2009, 12, 123–129.
- García-Rico, R.O.; Martín, J.F.; Fierro, F. Heterotrimeric Gα protein Pga1 from Penicillium chrysogenum triggers germination in response to carbon sources and affects negatively resistance to different stress conditions. Fungal Genet. Biol. 2011, 48, 641–649.
- Audhya, A.; Emr, S.D. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev. Cell. 2002, 2, 593–605.
- Audhya, A.; Foti, M.; Emr, S.D. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 2000, 11, 2673–2689.
- Voelker, D.R. Protein and lipid motifs regulate phosphatidylserine traffic in yeast. Biochem. Soc. Trans. 2005, 33, 1141–1145.
- Kishimoto, T.; Mioka, T.; Itoh, E.; Williams, D.E.; Andersen, R.J.; Tanaka, K. Phospholipid flippases and Sfk1 are essential for the retention of ergosterol in the plasma membrane. Mol. Biol. Cell. 2021, 32, 1374–1392.
- Chung, J.; Nakatsu, F.; Baskin, J.M.; De Camilli, P. Plasticity of PI4KIIIα interactions at the plasma membrane. EMBO Rep. 2015, 16, 312–320.
- Mioka, T.; Fujimura-Kamada, K.; Mizugaki, N.; Kishimoto, T.; Sano, T.; Nunome, H.; Williams, D.E.; Andersen, R.J.; Tanaka, K. Phospholipid flippases and Sfk1p, a novel regulator of phospholipid asymmetry, contribute to low permeability of the plasma membrane. Mol. Biol. Cell. 2018, 29, 1203–1218.
- Gil-Durán, C.; Rojas-Aedo, J.F.; Medina, E.; Vaca, I.; García-Rico, R.O.; Villagrán, S.; Levicán, G.; Chávez, R. The pcz1 gene, which encodes a Zn(II)2Cys6 protein, is involved in the control of growth, conidiation, and conidial germination in the filamentous fungus Penicillium roqueforti. PLoS ONE 2015, 10, e0120740.
- Torrent, C.; Gil-Durán, C.; Rojas-Aedo, J.F.; Medina, E.; Vaca, I.; Castro, P.; García-Rico, R.O.; Cotoras, M.; Mendoza, L.; Levicán, G.; et al. Role of sfk1 Gene in the filamentous fungus Penicillium roqueforti. Front. Microbiol. 2017, 8, 2424.
- Rojas-Aedo, J.F.; Gil-Durán, C.; Goity, A.; Vaca, I.; Levicán, G.; Larrondo, L.F.; Chávez, R. The developmental regulator Pcz1 affects the production of secondary metabolites in the filamentous fungus Penicillium roqueforti. Microbiol. Res. 2018, 212–213, 67–74.
- Ramamoorthy, V.; Dhingra, S.; Kincaid, A.; Shantappa, S.; Feng, X.; Calvo, A.M. The putative C2H2 transcription factor MtfA is a novel regulator of secondary metabolism and morphogenesis in Aspergillus nidulans. PLoS ONE 2013, 8, e74122.
- Zhuang, Z.; Lohmar, J.M.; Satterlee, T.; Cary, J.W.; Calvo, A.M. The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus. Toxins 2016, 8, 29.
- Sheridan, K.J.; Dolan, S.K.; Doyle, S. Endogenous cross-talk of fungal metabolites. Front. Microbiol. 2015, 5, 732.
- Bergmann, S.; Funk, A.N.; Scherlach, K.; Schroeckh, V.; Shelest, E.; Horn, U.; Hertweck, C.; Brakhage, A.A. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl. Environ Microbiol. 2010, 76, 8143–8149.
- Zhang, W.; Du, L.; Qu, Z.; Zhang, X.; Li, F.; Li, Z.; Qi, F.; Wang, X.; Jiang, Y.; Men, P.; et al. Compartmentalized biosynthesis of mycophenolic acid. Proc. Natl. Acad. Sci. USA 2019, 116, 13305–13310.
- Cepeda-García, C.; Domínguez-Santos, R.; García-Rico, R.O.; García-Estrada, C.; Cajiao, A.; Fierro, F.; Martín, J.F. Direct involvement of the CreA transcription factor in penicillin biosynthesis and expression of the pcbAB gene in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 2014, 98, 7113–7124.
- Chen, Y.; Li, B.; Xu, X.; Zhang, Z.; Tian, S. The pH-responsive PacC transcription factor plays pivotal roles in virulence and patulin biosynthesis in Penicillium expansum. Environ. Microbiol. 2018, 20, 4063–4078.
- Gente, S.; Poussereau, N.; Fèvre, M. Isolation and expression of a nitrogen regulatory gene, nmc, of Penicillium roqueforti. FEMS Microbiol. Lett. 1999, 175, 291–297.
More
