Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Arjan Kortholt and Version 2 by Conner Chen.
Mutations in the multidomain protein Leucine-rich-repeat kinase 2 (LRRK2) have been identified as a genetic risk factor for both sporadic and familial Parkinson’s disease (PD). LRRK2 has two enzymatic domains: a RocCOR tandem with GTPase activity and a kinase domain. In addition, LRRK2 has three N-terminal domains: ARM (Armadillo repeat), ANK (Ankyrin repeat), and LRR (Leucine-rich-repeat), and a C-terminal WD40 domain, all of which are involved in mediating protein–protein interactions (PPIs) and regulation of the LRRK2 catalytic core.
- Leucine-rich-repeat kinase 2
- Parkinson’s disease
- structures
1. From Bacterial Roco to Human Full-Length LRRK2 Structures
Structural studies on Leucine-rich-repeat kinase 2 (LRRK2) have been ongoing for almost two decades. However, the expression of the separate LRRK2 domains and full-length protein has been a major challenge for a long time. So far, only the crystal structures of the human LRRK2 Roc and WD40 domain have been resolved [1][2][16,17]. In addition, X-ray structures are available from homologous Roco family proteins. A major breakthrough in the field of structural biology in general and for determining the LRRK2 structure has been the recent developments in electron microscopy (EM). The possibilities related to stable and higher field emission EM, a robust vitrification apparatus, a volta phase plate, faster direct electron detector, and improved data analysing software have resulted in an increase in EM-based structure publications by over 70% in the last three years [3][4][5][18,19,20]. Using these techniques in combination with optimized purification protocols for full-length LRRK2 (flLRRK2) resulted in the first low-resolution negative-stain EM models of LRRK2 in 2016 [6][7][21,22]. More recently, the structures of a construct comprising Roc-COR-Kinase-WD40 domains (LRRK2RCKW) and flLRRK2 have been reported by Cryo-EM [8][9][10][23,24,25]. In addition, with Cryo-electron tomography (Cryo-ET), an in situ microtubule-bound full-length LRRK2 structure in cells was determined [11][26]. Another breakthrough in structural biology is the DeepMind-produced 3D protein structure prediction computational method, AlphaFold2, which makes it possible to predict reliable structures based on multiple sequence alignment and pair representation algorithms in silico [12][13][27,28].
2. Kinase Domain Structure
The LRRK2 kinase domain is considered the main output of the protein. It can phosphorylate a group of Rab GTPase family members to regulate several cellular pathways [14][33]. In addition, auto-phosphorylation of LRRK2 is also important for intramolecular regulation [15][16][34,35]. The protein kinase family was discovered by Krebs and Fisher during their study of glycogen phosphorylase in 1956 [17][36]. Kinase domains catalyze the transfer of γ-phosphate from ATP to a substrate. The kinase family is classified into three groups: Serine/Threonine kinase, tyrosine kinase, and atypical kinase. Based on the kinase domain sequence, LRRK2 belongs to the tyrosine kinase-like kinase (TKL) subfamily, a subfamily of Serine/Threonine kinase, whose members show sequence similarity to tyrosine kinases (TK) but lack TK-specific motifs [18][19][37,38].
A detailed structural map of the LRRK2 kinase domain first became available after the high-resolution LRRK2RCKW and flLRRK2 Cryo-EM structures were published in 2019 [8][9][23,24]. The structures revealed that LRRK2 has a typical kinase structure with a rich β-sheets-formed N-lobe and an α-helix-composed C-lobe. The α-helix in the C-lobe forms the activation segment, which consists of the DFGψ (DYGψ in LRRK2) motif—activation loop—P + 1 loop—APE motif—α-F, and the N-lobe is tightly anchored to the kinase C-lobe by the β4-αC loop [19][20][38,39]. However, due to flexibility, the structure of the activation segment has only been partly resolved in the LRRK2RCKW and flLRRK2 structures, while in the flLRRK2 structure, the DYGψ motif forms an unusual helix (Figure 12A). In the activation segment, the DYGψ motif works as a “brake” for LRRK2 conformational changes and activation: the Asp residue from the DFGψ motif binds with Mg2+ to indirectly interact with the oxygen of the β phosphate from ATP, and the P + 1 loop and APE motif provide the docking and interacting sites for substrates peptides, while the αF works as a scaffold that anchors the activation segment in a stable conformation in the kinase inactive state [21][40]. The two experimentally determined LRRK2 kinase structures both represent an inactive kinase conformation, with the activation segment collapsed on the protein interface in a locked position, thereby hindering substrate or ATP binding. In contrast, the predicted AlphaFold2 structure represents a DYGout active kinase conformation (Figure 12B). The conserved GxGxxG motif in the N-lobe forms most of the ATP binding pocket and is crucial for the stabilization of the phosphate and catalytic activity. Interestingly, the GxGxxG motif adopts a different conformation in the LRRK2RCKW and flLRRK2 (Figure 12A), which might be due to the different concentrations of ATP in the sample preparation.
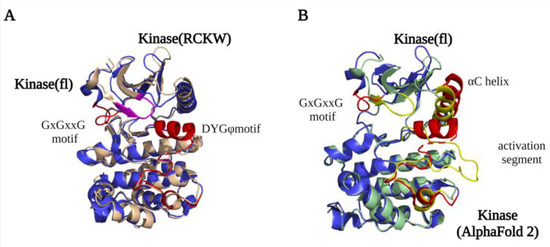
Figure 12. LRRK2 kinase domain structures. (A) An overlay of the kinase domain from LRRK2RCKW (PDB ID: 6VP7) (light yellow) and flLRRK2 (PDB ID: 7LHW) (blue). The main difference between the structures is the conformation of the GxGxxG motif (highlighted LRRK2RCKW in purple and flLRRK2 in red). (B) An overlay of the kinase domain from flLRRK2 (PDB ID: 7LHW) (blue) and AlphaFold2 model (green) structures. The GxGxxG motif, αC helix, and activation segments are highlighted in red (flLRRK2) and yellow (AlphaFold2), respectively.
By comparing the structures of the LRRK2 kinase domain with those of Braf (B-Raf Proto-Oncogene, Serine/Threonine kinase) and Src (Proto-oncogene tyrosine-protein kinase), the conserved Regulator spine (R-spine) and Catalytic spine (C-spine) were identified [22][41]. The C-spine has a substrate-capturing, nucleotide-binding, and phosphate transfer function, while the R-spine generally only assembles in kinase active state [23][42]. However, unexpectedly, the R-spine is also “visible” in the inactive LRRK2RCKW and flLRRK2 structures. The highly conserved Phe in the DFGψ motif is one of the four R-spine residues, and LRRK2 has, instead of a DFGψ motif, an unusual DYGψ motif. Interestingly, mutating DYGψ to DFGψ resulted in an LRRK2 hyperactive phenotype, suggesting that Y2018 is important for stabilizing a kinase inactive conformation [24][43]. The DYGψ motif thus serves as a conformational switch that regulates LRRK2 activation.
3. RocCOR Structure
The LRRK2 RocCOR domain has GTPase hydrolysis activity and, in addition, regulates LRRK2 dimerization [25][44]. As discussed in more detail below, dimerization of LRRK2 is important for GTPase and kinase activation.
The first LRRK2 Roc crystal structure reported by Deng et al. in 2008 showed that the GDP-bound Roc domain has a typical G domain fold with five α-helix and six β-strands [1][26][16,45]. The Roc domain has a highly conserved P-loop (G1), switch I motif (G2), and switch II motif (G3) that together are responsible for Mg2+ and nucleotide binding. The G4-G5 motifs are important for guanine specificity, and are only partly conserved in LRRK2 [27][28][29][46,47,48]. The structure shows that two Roc proteins assemble in a dimeric structure by the interaction of one Roc N-terminal with the C-terminal of the other protomer, thereby forming a pseudo-twofold [1][16]. However, this unusual dimeric structure is not observed in structures of homologous Roco family proteins nor in any of the high-resolution LRRK2 structures. Furthermore, both an N and C terminal extension of the Roc domain can disrupt the dimerization. Therefore, this structure is most likely caused by a crystallization artifact that was induced by the short flanking boundaries of the construct used [30][49]. Biochemical and structural characterization of the RocCOR domain of C. tepidum (Ct) Roco has revealed instead that the dimerization is mainly mediated by the C-terminal of the COR domain (CORB) [31][29]. The full-length CtRoco structure showed that in addition to the tight CORB interaction, the Roc domain is also involved in the dimeric interaction by Roc–Roc, Roc–CORA (N-terminal of the COR domain), and Roc–CORB interactions. These dimeric interfaces are mainly formed by the P-loop, switch II, and a highly conserved Roc dimerization loop between β4 and α3 [32]. An overlay of the CtRoco structures and a monomeric MbRocCORA of Methanosarcina barkeri (Mb) Roco2 structure show there are rearrangements of the P-loop, switch II, and CORA domain, suggesting that they might play an important role during the Roco G-protein cycle [33][31] (Figure 23A). Initially, it was proposed that Roco proteins form constitutive dimers; however, detailed biophysical and biochemical characterization of the CtRoco proteins revealed a monomer–dimer transition during the G-protein cycle [31][34][29,50]. The protein is mainly monomeric in the GTP state, while it forms dimers in the nucleotide-free and GDP-bound states [34][50]. Furthermore, an analogue of LRRK2 PD-associated mutation stabilizes the dimer and shows decreased GTPase activity [34][50]. Together, this shows that dimerization is an important step in the Roco activation mechanism.
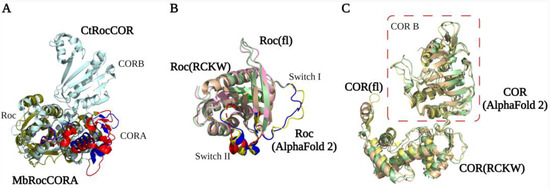
Figure 23. RocCOR tandem structures. (A) Alignment of the Mb (PDB ID: 4WNR) and Ct (PDB ID: 3DPU) RocCOR domains in dark green and light blue respectively. The rearrangement of P-loop, switch II, and CORA domain are indicated in red (MbRocCORA) and blue (CtRocCOR). (B,C) Alignment of the Roc/COR domain from LRRK2RCKW (PDB ID: 6VP7) (light yellow), flLRRK2 (PDB ID: 7LHW) (pink), and AlphaFold2 (green). The partly and totally missed switch I/II in LRRK2RCKW are highlighted in blue (flLRRK2), red (LRRK2RCKW), and yellow (AlphaFold2), respectively.
The LRRK2RCKW, flLRRK2, and predicted AlphaFold2 RocCOR structures all overlap well, with the exception of some differences observed for the Switch II in the LRRK2RCKW structure (Figure 23B). All three structures show that the COR domain consists of two subdomains that are connected by a flexible linker [8][9][23,24]. CORA consists of multiple α-helices and a short three-stranded antiparallel sheet (Figure 23C). The CORB sub-domain consists of four flanked helices, a central seven-stranded antiparallel sheet, and a hairpin motif. Consistent with the bacterial Roco structures, the CORB domain is the main dimerization interface in the flLRRK2 structure, which is formed by two partially overlapped β sheets [9][24]. In addition, the LRRK2RCKW single-particle and microtubule-associated structures revealed a WD40–WD40 domain interface [8][11][23,26]. How these COR–COR and WD40–WD40 interactions contribute to LRRK2 dimerization, and whether the nucleotide-dependent dimerization mechanism of CtRoco is conserved in LRRK2, need to be determined.
4. N-Terminal and C-Terminal Scaffold Structure
The LRRK2 N-terminal ARM, ANK (Ankyrin repeat), LRR, and C-terminal WD40 domains are tandem repeat domains that most likely are involved in PPIs with upstream and downstream regulators [35][36][37][51,52,53]. Recent data suggest that, in addition, these domains have intramolecular interactions that are involved in regulating LRRK2 activity. Despite numerous efforts, structural information about the N-terminal domains remains scarce. Even in the recently published flLRRK2 structure, most of the ARM domain is still not resolved, probably due to its flexibility [9][24] (Figure 34). The ARM domain that is visible in the structure also does not show the expected typical three-helical structure [35][51]. The AlphaFold2 model suggests that the LRRK2 ARM domain groove is shallower than in other typical ARM structures, but it does predict 14 repeats in the region of aa49-702 that form a typical three-helix patched solenoid structure, with the shortest helix, H1, perpendicular to the left two antiparallel helices. The H1 and H2 are located at the cylindrical outer surface and are predicted to, together with H3, mediate most of the PPIs [38][54].
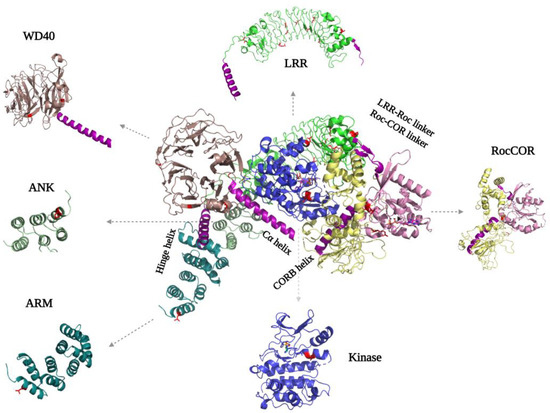
Figure 34. FlLRRK2 (PDB ID: 7LHW) with the separated domain’s structure on the sides. The helixes that are important for interdomain mediation are highlighted in purple. The PD mutations are indicated in both the full-length structure and separate domains in red, where they are located either in intradomain or interdomain interfaces.
For the ANK domain, it was initially predicted that it consists of seven repeats from aa679-902, while a later study proposed it consists of five repeats from aa690 to aa860 [35][39][51,55]. However, both the flLRRK2 and AlphaFold2 structures show that the ANK domain only comprises three typical antiparallel helix repeats (aa705-795) that are connected by a hairpin (Figure 34). The LRRK2 ANK domain forms a typical L-shaped structure with the extended hairpin exposed to the outside to mediate PPIs [40][41][42][56,57,58]. The structures also revealed that the predicted C-terminal segment of the ANK domain (aa795-860) is actually part of the LRR domain, where it functions as a hinge helix that connects the LRR domain with the ANK, ARM, and WD40 domains [9][24].
The LRR domain of flLRRK2 is folded into a 14 + 1 repeat horse-shaped structure [9][24]. The beta-strand is located at the concave surface, and it is connected with the convex side α-helix by a loop [9][24]. After the last strand-helix repeat at the C-terminus of the LRR domain, there is a short linker that crosses the Roc–CORA linker and forms a connection with the Roc domain (Figure 34). In the CtRoco protein, this linker between the LRR and Roc domain is around 40 residues in length and includes a PLxxPPPE motif that is conserved in prokaryotic Roco proteins [31][29]. This linker in CtRoco was shown to be important for the stability of the Roc–COR construct protein, and the crystal structure showed direct interaction of the linker with the Roc domain switch II motif [31][32][29,32]. However, in both the flLRRK2 and AlphaFold2 structures, the linker and LRR domain are far away from the Roc catalytic core.
The crystal structure of the dimeric WD40 domain published by Zhang et al. in 2019 revealed a typical seven blade ring-like structure, each consisting of four anti-parallel β-strands [2][17] (Figure 34). The WD40–WD40 dimeric interface is formed by the circumference of blades V, IV, and VI, rather than by the popular blade top surfaces. Consistently, this same WD40 dimer interface is present in the microtubule-bound LRRK2 in situ structure and the trimeric LRRK2RCKW EM structure [8][11][23,26]. Interestingly, the extended LRRK2RCKW, flLRRK2, and AlphaFold2 structures also show that the C-terminal helix of the WD40 domain directly interacts with the CORB and kinase domain, suggesting that the WD40 domain might regulate the enzymatic activity of LRRK2 [9][24] (Figure 34).
