Crude oil contamination is an emerging environmental concern on a global scale due to inadvertent oil leakage [1]. Oil spillage and discharges frequently happen as a consequence of explosion incidents during oilfield drilling; leakage from oil and gas pipelines and reservoirs, fuel tankers, and well waxing; and during overhauls of refineries and petrochemical manufacturing equipment. Crude oil and its products damage water and air quality and also reduce soil fertility. Crude oil can harm plants by clogging soil pores, reducing soil aeration and water permeability, which can have both ecological and toxicological effects and disrupt the soil’s natural structure. Aside from these environmental concerns, one of the most serious consequences of such anthropogenic emissions is current global climate change. Crude oil contamination not only impacts different ecosystems but also influences global socio-economic status.
- crude oil contamination
- bioremediation
- bacteria
- biodegradation
- metabolic engineering
1. Introduction
With all the shortcomings of other techniques of remediation, the complete or partially effective removal of crude oil is still a challenge [43,44,45][1][2][3]. The microbial degradation of complex and diverse crude oil molecules requires specific enzymes for a specific class of compounds [46,47][4][5]. Depending on the bacterial species and community, the resulting catabolites are either utilised by the bacteria themselves or released into the environment for further degradation by the other bacteria in the community [48][6]. The sole determining factor critical to the degradation is the survival of the bacteria in a medium with high crude oil contamination [49][7]. Due to their capacity to degrade a variety of crude oil components, bacteria are considered to be the most effective degraders of crude oil [50,51,52][8][9][10]. According to the research by El-Liethy et al. [53][11] the rate of crude oil degradation by the bacterial strain Enterobacter hormaechei was 0.6% in a minimal medium, but it accelerated to 70.7% when the medium was biostimulated with peptone. A bioremediation experiment carried out with biosurfactant-producing Pseudomonas aeruginosa showed a maximum of 68.3% degradation of n-hexadecane in 60 days [54][12]. Alkanes with different molecular chains may be degraded by the Acinetobacter strain DSM17874 using degrading genes, such as alkB and AlmA [55][13]. Pseudomonas nitroreducens efficiently degraded 70% of paraffinic contaminants in a short period of 10 days [56][14]. However, a consortium can attain better degradation of crude oil by the availability of a broader range of enzymes acting upon crude oil compounds, as well as the co-existence of multiple bacteria that assist in the formation of higher metabolic networks, which can interact with the persistent compounds and carry out the process of biodegradation [52,57][10][15]. A microbial consortium consisting of Actinotalea ferrariae, Arthrobacter ginsengisoli, Dietzia cinnamea, Dietzia papillomatosis, and Pseudomonas songnensis showed a degradation of crude oil ranging from 73.6 to 69.3% in soil with 1–10% crude oil contamination [58][16].
2. Regulatory Factors Involve in Microbial Degradation of Crude Oil
Microbes are primarily responsible for the biodegradation of crude oil in the contaminated environment; however, several biotic and abiotic factors also influence the efficacy of bioremediation approaches, such as soil oxygen content, soil type, pH, temperature, nutrient availability, water content, and the concentration of crude oil and its bioavailability in the existing environment [94][35]. The bioavailability of crude oil components in a soil environment is an essential factor in regulating biodegradation rate [95][36]. Though the increase in the concentration of crude oil negatively affects the degradation rate, the degradation of aromatic compounds and linear hydrocarbons occurs discretely [96][37]. After the rapid initial rate of crude oil degradation, the residue becomes partially diffused to the surrounding solid surfaces, reducing its bioavailability, phosphorus and nitrogen contents and degradation rate at later stages [97][38]. The improper disposal of crude oil with high concentrations of volatile hydrocarbons and sludge (15–20%) can harm the microbial population and hinder the biodegradation of the oil [98,99][39][40]. Temperature has multiple effects in the bioremediation process, as it influences the rate at which the crude oil is degraded by the microorganisms, affecting the composition of the whole soil microbiome [100][41]. The chemical and physical properties of crude oil are also affected by temperature variations. The soil–water partition coefficient decreases with increasing temperature in soil with high moisture content, and contaminant dissolution may occur [101][42]. Though the ability of microorganisms to degrade crude oil is negatively affected in high-pH environments [102][43], the pH of the soil plays a major role in the degradation of crude oil by the bacteria. It has been observed that pH 7 is optimum for the significant degradation of PAHs and other n-alkanes (C-7 to C-25). However, bacterial strains such as Bacillus subtilis BL-27 showed a wide tolerance to pH values ranging from 4 to 10 during the degradation of crude oil [103][44]. Dubinsky et al. [104][45] observed that during unmitigated flow, n-alkane and cycloalkane availability were higher, which probably helped alkane-degrading bacteria flourish. According to metagenomic and meta-transcriptomic analyses carried out on a subset of samples obtained during uncontrolled flow, alkane degradation was shown to be the predominant hydrocarbon-degrading pathway expressed when Oceanospirillaceae and Pseudomonas were prevalent in the community. Numerous investigations demonstrated that oxygen loss causes biodegradation activities in soils and marine sediments to decrease drastically [105,106,107][46][47][48]. There are studies that suggest the estimated oxygen requirements for aerobic hydrocarbon degradation. As 3kg O2 is required for every kg of petroleum contaminants, 8.6 moles of oxygen are required for every mole of diesel to be degraded [108][49]. Even though a variety of microbial communities typically contribute to the in-situ breakdown of alkane mixtures under varying soil circumstances, many soils show a similar pattern in the types of microorganisms that respond to alkane disturbance [109][50]. For appropriate bioremediation experiments, factors such as expense, duration, human resource, and targets must all be considered.3. Classical Metabolic Pathways Involved in Degradation of Total Petroleum Hydrocarbons
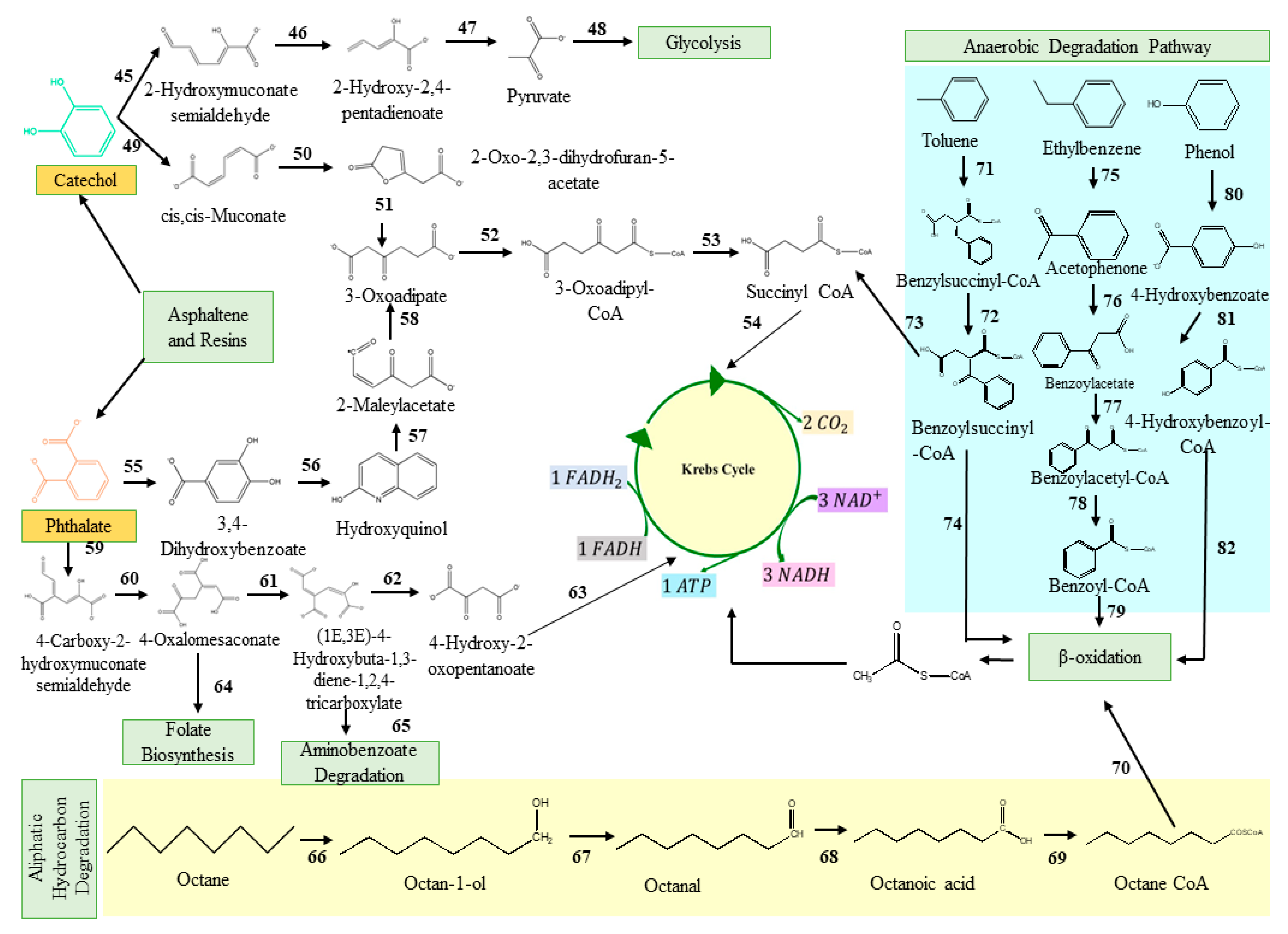
Hydrocarbonoclastic bacterial species degrade PAHs due to their adaptability, vigour, diversity, and capacity to generate less hazardous metabolic intermediates. Microbes use the energy from the breakdown of hydrocarbons ranging from simple alkanes to PAHs [110][51]. Oxygen is necessary for the ring hydroxylation, ring cleavage, and final electron uptake in the aerobic breakdown of PAHs by bacteria. However, reductive processes are the basis of anaerobic PAH consumption [95,111][36][52]. High-molecular-weight PAHs are harder to break down because their fused aromatic structures make them very thermodynamically stable, water-repellent, and less bioavailable. Pyrene mineralisation can take place either at the C-1 and C-2 positions or at the C-4 and C-5 positions of the aromatic ring due to the action of the dioxygenase. Following that, the ring aromatisation of the dihydrodiols and the ring cleavage dioxygenase led to the synthesis of phenanthrene dicarboxylate, which is then decarboxylated to generate carboxylate. The phenanthrene dicarboxylate is eventually converted into phenanthrene carboxylate. At this stage, a cis-3,4-dihydroxyphenanthrene-4-carboxylate is formed by a deoxygenation process. This cis-3,4-dihydroxyphenanthrene-4-carboxylate then rearomatizes to form dihydroxyphenanthrene, which is then metabolised to yield hydroxynaphthoate. In addition, the pyrene breakdown occurs via the phthalate pathway as well [112,113][53][54]. The phthalate reaction forms several intermediates, including 3,4-dihydroxybenzoate and carboxyhydroxymuconate semialdehyde, both of which eventually feed into the Krebs cycle via the multiple reaction steps. Phenanthrene is a three-ringed compound whose degradation has been reported in both Gram-positive and Gram-negative microbes [114,115][55][56]. Most of the intermediates of phenanthrene degradation are shared with the pyrene degradation pathway. Naphthalene, toluene, and anthracene have a comparatively low molecular weight and constitute most petroleum hydrocarbons [116][57]. Ring hydroxylating genes, such as nagAc, phnAc, nahAc, nidA, and pdoA, are target genes for naphthalene breakdown. In the first stage, an oxidoreductase inserts two oxygen atoms into the naphthalene rings to generate dihydronaphthalene. The following step involves the dehydrogenase enzyme, followed by ring cleavage and oxidation [117][58]. The dioxygenase facilitates the cleavage of catechol and proceeds via various pathways, ultimately leading to the formation of succinyl-CoA and entering the TCA cycle [118][59]. Figure 1 represents the classical pathways for the aerobic biodegradation of various hydrocarbons, such as pyrene, anthracene, phenanthrene, and naphthalene (reconstructed from the KEGG database). Microorganisms synthesised several catabolic enzymes, such as naphthalene 1,2-dioxygenase, oxidoreductases, pyrene dioxygenase, aldehyde dehydrogenase, and many others, in every step of the degradation process. The enzymes involved in the different steps of the degradation process are also given in Figure 1.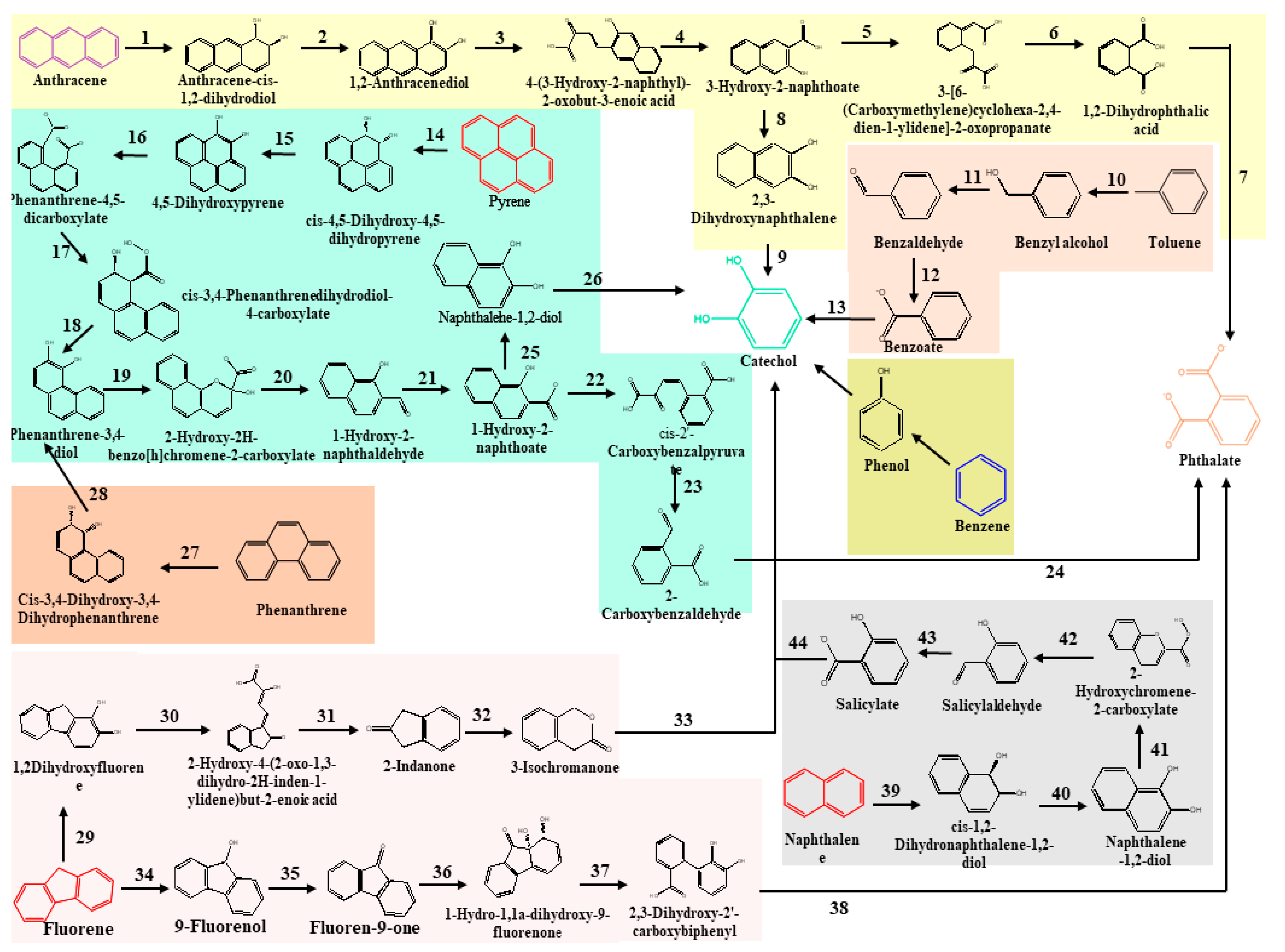
References
- Rahman, K.S.M.; Thahira-Rahman, J.; Lakshmanaperumalsamy, P.; Banat, I.M. Towards efficient crude oil degradation by a mixed bacterial consortium. Bioresour. Technol. 2002, 85, 257–261.
- Das, K.; Mukherjee, A.K. Crude petroleum-oil biodegradation efficiency of Bacillus subtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresour. Technol. 2007, 98, 1339–1345.
- Zucchi, M.; Angiolini, L.; Borin, S.; Brusetti, L.; Dietrich, N.; Gigliotti, C.; Barbieri, P.; Sorlini, C.; Daffonchio, D. Response of bacterial community during bioremediation of an oil-polluted soil. J. Appl. Microbiol. 2003, 94, 248–257.
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A Comprehensive Review of Aliphatic Hydrocarbon Biodegradation by Bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699.
- Megharaj, M.; Ramakrishnan, B.; Venkateswarlu, K.; Sethunathan, N.; Naidu, R. Bioremediation approaches for organic pollutants: A critical perspective. Environ. Int. 2011, 37, 1362–1375.
- Gargouri, B.; Karray, F.; Mhiri, N.; Aloui, F.; Sayadi, S. Bioremediation of petroleum hydrocarbons-contaminated soil by bacterial consortium isolated from an industrial wastewater treatment plant. J. Chem. Technol. Biotechnol. 2014, 89, 978–987.
- Roy, A.S.; Baruah, R.; Borah, M.; Singh, A.K.; Boruah, H.P.D.; Saikia, N.; Deka, M.; Dutta, N.; Bora, T.C. Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int. Biodeterior. Biodegrad. 2014, 94, 79–89.
- Lee, H.; Yun, S.Y.; Jang, S.; Kim, G.H.; Kim, J.J. Bioremediation of polycyclic aromatic hydrocarbons in creosote-contaminated soil by Peniophora incarnata KUC8836. Bioremed. J. 2015, 19, 1–8.
- Hesnawi, R.M.; Adbeib, M.M. Effect of Nutrient Source on Indigenous Biodegradation of Diesel Fuel Contaminated Soil. APCBEE Procedia 2013, 5, 557–561.
- Gurav, R.; Lyu, H.; Ma, J.; Tang, J.; Liu, Q. Degradation of n-alkanes and PAHs from the heavy crude oil using salt-tolerant bacterial consortia and analysis of their catabolic genes. Environ. Sci. Pollut. Res. 2017, 24, 11392–11403.
- El-Liethy, M.A.; El-Noubi, M.M.; Abia, A.L.K.; El-Malky, M.G.; Hashem, A.I.; El-Taweel, G.E. Eco-friendly bioremediation approach for crude oil-polluted soils using a novel and biostimulated Enterobacter hormaechei ODB H32 strain. Int. J. Environ. Sci. Technol. 2022, 19, 10577–10588.
- Hajieghrari, M.; Hejazi, P. Enhanced biodegradation of n-Hexadecane in solid-phase of soil by employing immobilized Pseudomonas Aeruginosa on size-optimized coconut fibers. J. Hazard. Mater. 2020, 389, 122134.
- Throne-Holst, M.; Wentzel, A.; Ellingsen, T.E.; Kotlar, H.K.; Zotchev, S.B. Identification of novel genes involved in long-chain n-alkane degradation by Acinetobacter sp. strain DSM 17874. Appl. Environ. Microbiol. 2007, 73, 3327–3332.
- Okoye, A.U.; Chikere, C.B.; Okpokwasili, G.C. Characterization of potential paraffin wax removing bacteria for sustainable biotechnological application. In Proceedings of the Society of Petroleum Engineers—SPE Nigeria Annual International Conference and Exhibition 2019, NAIC 2019, Lagos, Nigeria, 6 August 2019.
- Rahman, R.N.Z.R.A.; Latip, W.; Adlan, N.A.; Sabri, S.; Ali, M.S.M. Bacteria consortia enhanced hydrocarbon degradation of waxy crude oil. Arch. Microbiol. 2022, 204, 701.
- Ali, N.; Khanafer, M.; Al-Awadhi, H. Indigenous oil-degrading bacteria more efficient in soil bioremediation than microbial consortium and active even in super oil-saturated soils. Front. Microbiol. 2022, 13, 2840.
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 2018, 9, 2885.
- Lakshmi, G.; Kishore Babu, A. Isolation and Study of Biodegradation Capability of Hydrocarbonoclastic Bacteria from Industrial Waste Lubrication Oil Contaminated Sites. Int. J. Curr. Microbiol. Appl. Sci. 2021, 10, 163–172.
- Xu, J.; Zhang, Q.; Li, D.; Du, J.; Wang, C.; Qin, J. Rapid degradation of long-chain crude oil in soil by indigenous bacteria using fermented food waste supernatant. Waste Manag. 2019, 85, 361–373.
- Mohanty, G.; Mukherji, S. Biodegradation rate of diesel range n-alkanes by bacterial cultures Exiguobacterium aurantiacum and Burkholderia cepacia. Int. Biodeterior. Biodegrad. 2008, 61, 240–250.
- Brito, E.M.S.; Guyoneaud, R.; Goñi-Urriza, M.; Ranchou-Peyruse, A.; Verbaere, A.; Crapez, M.A.; Wasserman, J.C.A.; Duran, R. Characterization of hydrocarbonoclastic bacterial communities from mangrove sediments in Guanabara Bay, Brazil. Res. Microbiol. 2006, 157, 752–762.
- Thangarajan, R.; Adetutu, E.M.; Moore, R.B.; Ogunbanwo, S.T.; Ball, A.S. Comparison between different bio-treatments of a hydrocarbon contaminated soil from a landfill site. Afr. J. Biotechnol. 2011, 10, 15151–15162.
- Bociu, I.; Shin, B.; Wells, W.; Kostka, J.E.; Konstantinidis, K.T.; Huettel, M. Decomposition of sediment-oil-agglomerates in a Gulf of Mexico sandy beach. Sci. Rep. 2019, 9, 10071.
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 2014, 98, 4781–4794.
- Zhou, L.; Li, H.; Zhang, Y.; Han, S.; Xu, H. Sphingomonas from petroleum-contaminated soils in Shenfu, China and their PAHs degradation abilities. Braz. J. Microbiol. 2016, 47, 271–278.
- Ganesh Kumar, A.; Mathew, N.C.; Sujitha, K.; Kirubagaran, R.; Dharani, G. Genome analysis of deep sea piezotolerant Nesiotobacter exalbescens COD22 and toluene degradation studies under high pressure condition. Sci. Rep. 2019, 9, 18724.
- Zhang, X.; Kong, D.; Liu, X.; Xie, H.; Lou, X.; Zeng, C. Combined microbial degradation of crude oil under alkaline conditions by Acinetobacter baumannii and Talaromyces sp. Chemosphere 2021, 273, 129666.
- Miglani, R.; Parveen, N.; Kumar, A.; Ansari, M.A.; Khanna, S.; Rawat, G.; Panda, A.K.; Bisht, S.S.; Upadhyay, J.; Ansari, M.N. Degradation of xenobiotic pollutants: An environmentally sustainable approach. Metabolites 2022, 12, 818.
- Turner, J.T. Zooplankton faecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog. Oceanogr. 2015, 130, 205–248.
- Hazen, T.C.; Prince, R.C.; Mahmoudi, N. Marine Oil Biodegradation. Environ. Sci. Technol. 2016, 50, 2121–2129.
- Dyksterhouse, S.E.; Gray, J.P.; Herwig, R.P.; Lara, J.C.; Staley, J.T. Cycloclasticus pugetii gen-nov, sp-nov, an aromatic hydrocarbon-degrading bacterium from marine-sediments. Int. J. Syst. Bacteriol. 1995, 45, 116−123.
- Bruns, A.; Berthe-Corti, L. Fundibacter jadensis gen. nov., sp. nov., a new slightly halophilic bacterium, isolated from intertidal sediment. Int. J. Syst. Bacteriol. 1999, 49, 441−448.
- Yakimov, M.M.; Giuliano, L.; Gentile, G.; Crisafi, E.; Chernikova, T.N.; Abraham, W.R.; Lunsdorf, H.; Timmis, K.N.; Golyshin, P.N. Oleispira antarctica gen. nov., sp nov., a novel hydrocarbonoclastic marine bacterium isolated from Antarctic coastal sea water. Int. J. Syst. Evol. Microbiol. 2003, 53, 779−785.
- Wang, B.; Lai, Q.; Cui, Z.; Tan, T.; Shao, Z. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. 2008, 10, 1948–1963.
- Das, N.; Bhuyan, B.; Pandey, P. Correlation of soil microbiome with crude oil contamination drives detection of hydrocarbon degrading genes which are independent to quantity and type of contaminants. Environ. Res. 2022, 215 Pt 1, 114185.
- Meckenstock, R.U.; Safinowski, M.; Griebler, C. Anaerobic degradation of polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 2004, 49, 27–36.
- Varjani, S.; Thaker, M.; Upasani, V. Optimization of growth conditions of native hydrocarbon utilizing bacterial consortium “HUBC” obtained from petroleum pollutant contaminated sites. Indian J. Appl. Res. 2014, 4, 474–476.
- Shafiee, P.; SHOJA, A.S.; Charkhabi, A.H. Biodegradation of polycyclic aromatic hydrocarbons by aerobic mixed bacterial culture isolated from hydrocarbon polluted soils. Iran. J. Chem. Chem. Eng. 2006, 25, 73–78.
- Edet, U.O.; Anika, O.C.; Umoren, E. Toxicity Profile of Crude Oil on Degrading Bacterial Isolates. Int. J. Sci. Eng. Res. 2019, 10, 1131–1135.
- Obi, L.U.; Atagana, H.I.; Adeleke, R.A. Isolation and characterisation of crude oil sludge degrading bacteria. SpringerPlus 2016, 5, 1946.
- Ribicic, D.; McFarlin, K.M.; Netzer, R.; Brakstad, O.G.; Winkler, A.; Throne-Holst, M.; Størseth, T.R. Oil type and temperature dependent biodegradation dynamics—Combining chemical and microbial community data through multivariate analysis. BMC Microbiol. 2018, 18, 83.
- Devatha, C.P.; Vishnu Vishal, A.; Purna Chandra Rao, J. Investigation of physical and chemical characteristics on soil due to crude oil contamination and its remediation. Appl. Water Sci. 2019, 9, 89.
- Al-Hawash, A.B.; Dragh, M.A.; Li, S.; Alhujaily, A.; Abbood, H.A.; Zhang, X.; Ma, F. Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquat. Res. 2018, 44, 71–76.
- Wang, D.; Lin, J.; Lin, J.; Wang, W.; Li, S. Biodegradation of petroleum hydrocarbons by bacillus subtilis BL-27, a strain with weak hydrophobicity. Molecules 2019, 24, 3021.
- Dubinsky, E.A.; Conrad, M.E.; Chakraborty, R.; Bill, M.; Borglin, S.E.; Hollibaugh, J.T.; Mason, O.U.; Piceno, Y.M.; Reid, F.C.; Stringfellow, W.T.; et al. Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the Gulf of Mexico. Environ. Sci. Technol. 2013, 47, 10860–10867.
- Atlas, R.M.; Stoeckel, D.M.; Faith, S.A.; Minard-Smith, A.; Thorn, J.R.; Benotti, M.J. Oil Biodegradation and oil-degrading microbial populations in marsh sediments impacted by oil from the Deepwater Horizon well blowout. Environ. Sci. Technol. 2015, 49, 8356–8366.
- Alegbeleye, O.O.; Opeolu, B.O.; Jackson, V.A. Polycyclic aromatic hydrocarbons: A critical review of environmental occurrence and bioremediation. Environ. Manag. 2017, 60, 758–783.
- Kliem, S.; Kreutzbruck, M.; Bonten, C. Review on the biological degradation of polymers in various environments. Materials 2020, 13, 4586.
- Fragkou, E.; Antoniou, E.; Daliakopoulos, I.; Manios, T.; Theodorakopoulou, M.; Kalogerakis, N. In situ aerobic bioremediation of sediments polluted with petroleum hydrocarbons: A critical review. J. Mar. Sci. Eng. 2021, 9, 1003.
- Hamamura, N.; Olson, S.H.; Ward, D.M.; Inskeep, W.P. Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl. Environ. Microbiol. 2006, 72, 6316–6324.
- Kumari, S.; Regar, R.K.; Manickam, N. Improved polycyclic aromatic hydrocarbon degradation in a crude oil by individual and a consortium of bacteria. Bioresour. Technol. 2018, 254, 174–179.
- Baboshin, M.; Golovleva, L. Aerobic bacterial degradation of polycyclic aromatic hydrocarbons (PAHs) and its kinetic aspects. Microbiology 2012, 81, 639–650.
- Sun, S.; Wang, H.; Chen, Y.; Lou, J.; Wu, L.; Xu, J. Salicylate and phthalate pathways contributed differently on phenanthrene and pyrene degradations in Mycobacterium sp. WY10. J. Hazard. Mater. 2019, 364, 509–518.
- Vaidya, S.; Jain, K.; Madamwar, D. Metabolism of pyrene through phthalic acid pathway by enriched bacterial consortium composed of Pseudomonas, Burkholderia, and Rhodococcus (PBR). 3 Biotech 2017, 7, 29.
- Moody, J.D.; Freeman, J.P.; Doerge, D.R.; Cerniglia, C.E. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 2001, 67, 1476–1483.
- Zhao, H.-P.; Wu, Q.-S.; Wang, L.; Zhao, X.-T.; Gao, H.-W. Degradation of phenanthrene by bacterial strain isolated from soil in oil refinery fields in Shanghai China. J. Hazard. Mater. 2009, 164, 863–869.
- Peng, R.H.; Xiong, A.S.; Xue, Y.; Fu, X.Y.; Gao, F.; Zhao, W.; Tian, Y.S.; Yao, Q.H. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008, 32, 927–955.
- Barnsley, E.A. Naphthalene metabolism by pseudomonads: The oxidation of 1, 2-dihydroxynaphthalene to 2-hydroxychromene-2-carboxylic acid and the formation of 2′-hydroxybenzalpyruvate. Biochem. Biophys. Res. Commun. 1976, 72, 1116–1121.
- Anokhina, T.; Esikova, T.; Gafarov, A.; Polivtseva, V.; Baskunov, B.; Solyanikova, I. Alternative naphthalene metabolic pathway includes formation of ortho-phthalic acid and cinnamic acid derivatives in the Rhodococcus opacus strain 3D. Biochemistry 2020, 85, 355–368.
- Ahmed, H.M.; Kamal, M.S.; Al-Harthi, M. Polymeric and low molecular weight shale inhibitors: A review. Fuel 2019, 251, 187–217.
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490.
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58.
- Wang, W.; Shao, Z. Enzymes and genes involved in aerobic alkane degradation. Front. Microbiol. 2013, 4, 116.
- Zuo, P.; Qu, S.; Shen, W. Asphaltenes: Separations, structural analysis and applications. J. Energy Chem. 2019, 34, 186–207.
- Parra-Barraza, H.; Hernández-Montiel, D.; Lizardi, J.; Hernández, J.; Urbina, R.H.; Valdez, M.A. The zeta potential and surface properties of asphaltenes obtained with different crude oil/n-heptane proportions. Fuel 2003, 82, 869–874.
- Foght, J. Anaerobic biodegradation of aromatic hydrocarbons: Pathways and prospects. Microb. Physiol. 2008, 15, 93–120.
