Oxidative stress (OS) is defined as an imbalance between free radicals biogenesis and the cell antioxidant capacity to eliminate them. In neurodegenerative diseases, OS play a central role altering mitochondrial metabolism, protein synthesis, and inducing cellular malfunctioning. Most part of the human genome encodes for non-coding protein genes, which are transcribed into non-coding RNA (ncRNA). Most of these ncRNAs are involved transcriptional and post-transcriptional regulation of gene and their deregulation has been linked to diverse neurodegenerative disorders. In this review we compiled most recent evidences reporting a role of main types of ncRNAs in the regulation and management of oxidative stress in Alzheimer's disease, Parkinson's disease, Huntington's disease and Amyotrophic Lateral Sclerosis.
- oxidative stress
- neurodegeneration
- ncRNA
- miRNA
- tRNA fragments
- lncRNA
- circRNA
1. Introduction
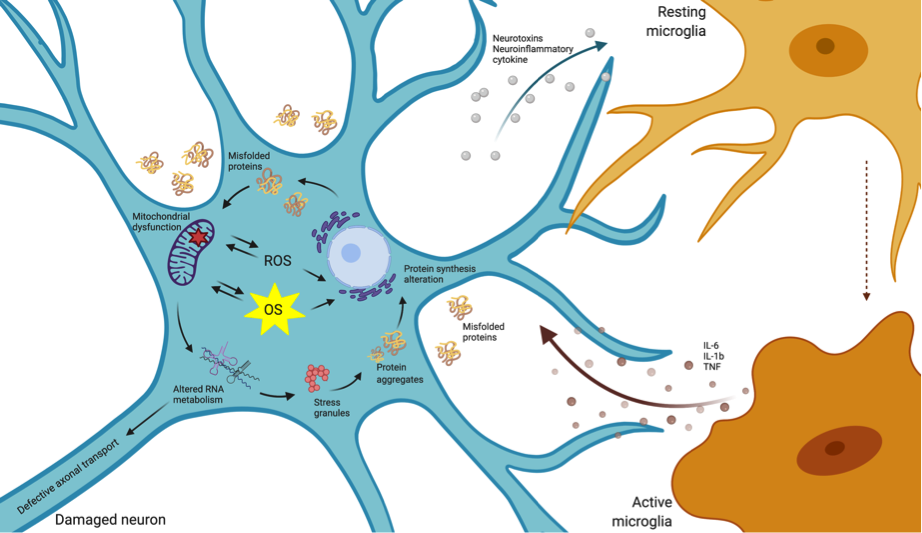
Neurodegenerative disorders are a heterogeneous class of diseases characterized by a slow, progressive neuronal dysfunction and loss. The etiology of this class of diseases has not been fully elucidated, with highly complex and multifactorial causes participating in disease onset and progression. Nevertheless, several lines of evidence suggest that oxidative stress (OS) has a central role in this type of disorders. OS is the result of an imbalance between the increase in reactive oxygen species (ROS) and the production of antioxidants to eliminate them. A variety of mechanisms involving oxidative damage have been described in neurodegenerative conditions, including mitochondrial dysfunction, the oxidation of nucleic acids, proteins and lipids, activation of glial cells, apoptosis, cytokines production and inflammatory responses (Figure 1). The identification of molecular modifiers of the endogenous antioxidant activity is key to boost novel therapeutic designs [1].
The advent of high-throughput sequencing technologies revealed that the human genome is pervasively transcribed; with a large proportion corresponding to non-coding RNA (ncRNA) transcripts, the most abundant class of RNAs in cells[2][3]. ncRNAs have been classified according to their length and activity[4] with many classes revealed as essential regulators of gene expression through a variety of mechanisms[5].
ncRNAs are especially abundant and diverse in the central nervous system (CNS), showing a highly specific and dynamic temporal and spatial expression pattern that guides neurodevelopmental processes, including neural stem cell proliferation and differentiation[6][7]. In addition to their role in developmental processes, ncRNAs are key for the maintenance and normal functioning of adult, post-mitotic neurons. Most studies have been focused on microRNAs (miRNAs), the best-known class of ncRNAs. Although the roles for a progressively increasing number of ncRNAs are continuously being uncovered, the biological functions of the vast majority remain unknown.
While the precise expression levels of ncRNAs are key for the normal function of the CNS, the deregulation of ncRNA pathways have been linked to human disease, including neurodegenerative conditions[8][9][10][11]. In the present manuscript, we will provide an overview on the functional association between ncRNAs and the response to OS, a major hallmark in neurodegenerative disorders, and we will review the OS–ncRNA axis in major neurodegenerative conditions.
2. Regulatory Non-Coding RNAs in the Normal Function of the Nervous System
NcRNAs are classified as housekeeping ncRNAs and regulatory ncRNAs. Housekeeping ncRNA are highly expressed constitutively and ubiquitously and play crucial roles in routine cell maintenance; they include transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). Regulatory ncRNAs, which are the focus of the present review, are characterized by a highly regulated and specific expression pattern, by their activity as transcriptional and post-transcriptional gene expression modulators and their high sensitivity to cell needs[12]. Regulatory ncRNAs can be classified based on their length as long non-coding RNAs (lncRNAs; >200 nucleotides) and short/small non-coding RNAs (sncRNAs; <200 nucleotides, usually between 18 and 40 nt)[11]. Discoveries over the last 40 years have elucidated diverse functions for ncRNAs involving the regulation of mRNA expression stability and translation, including splicing regulation, chromosome maintenance and segregation, and even the transmission of regulatory signals.
In particular, RNA biology is of paramount significance in the CNS. The development of CNS and brain morphogenesis is a very complex spatiotemporal-organized process and requires an exquisite regulation of stem cell proliferation and differentiation. The brain is the organ showing the highest diversity of regulatory ncRNAs [7][13][14], and their dynamic expression and activity underlies CNS complex functions including neurodevelopment, synapse function, cell polarity and maturation, and the responses to intra- and extracellular stimuli [6]. In this section, we outline the role of the main classes of regulatory ncRNAs in the development and correct function of the nervous system.
2.1. microRNAs
microRNAs (miRNAs, ~21 nt), the most studied class of regulatory ncRNAs, were discovered in 1993 in C. elegans [15]. They are one of the main negative post-transcriptional regulators of gene expression. This regulation is achieved by the direct, incomplete, base pair targeting of mRNAs, which results in the recruitment of diverse RNA-decay factors, or in translation repression.
Although miRNA expression and correct biogenesis is important in the whole organism, they appear as indispensable molecules for CNS development and adult brain function[16][17]. In fact, the depletion of miRNA biogenesis-related proteins Dicer-1 or DGCR8, strongly affects embryonic and neural development[16][18][19][20]. Chmielarz and colleagues recently reported that the specific removal of Dicer in dopaminergic neurons leads to the progressive degeneration of these cells in mice striatum and consequent locomotor affectation[21]. miRNAs also orchestrate neural signaling processes including neurotransmitter release and dendrites and spines generation. Specifically, diverse evidence points to miRNAs as important regulators of pre-synaptic and post-synaptic responses.
miRNAs have likewise been studied in glial cells. Several works on murine cells and animal models have reported the role of specific miRNAs in gliogenesis[21] and glial differentiation[22]. Additionally, other miRNAs have been implicated in cerebral immune response through the regulation of glial cell proliferation and microglial polarization[23][24], as reviewed by Guo et al.[25].
2.2. tRNA-Derived Fragments
tRNAs-derived fragments (tRFs) (<40 nt) are bioactive molecules classified according to their size and the part of the mature tRNA from where they derive, as previously reviewed in Kumar et al.[26]. tRFs can be constitutively produced and/or induced as a result of cellular stress response pathways. Diverse nucleases are responsible for the biogenesis of tRFs in vertebrates including RNase Z, Dicer and angiogenin (ANG). Under stress conditions, ANG produces a subtype of tRNA halves (tRNA-derived stress-induced RNAs or tiRNAs)[27], and Dicer is involved in the biogenesis of tRFS derived from the D loop or the 3′-end of mature tRNAs[28]. However, the role of Dicer in tRF biogenesis may be limited, as its depletion has little impact on tRF levels in multiple model organisms [29].
tRFs are highly abundant in the brain, where they accumulate during ageing[30][31], pointing these species as risk factors in neurodegenerative conditions. Furthermore, tRF expression dynamics have been associated to stem cell differentiation[32], denoting that the biogenesis of specific tRFs is an indicator of cell status and participates in biological processes associated to particular physiological or pathological conditions.
Although tRFs’ heterogenous functions are not entirely understood, they regulate mRNA processing and translation, controlling processes such as cell growth and differentiation[33]. tRFs may could work as miRNA inhibiting gene expression[34][35], and they can also regulate gene expression by competitively binding to AGO proteins, thus affecting the silencing activity on target genes[36]. Furthermore, tRFs repress translation through binding to ribosomal factors, emphasizing their implication in multiple layers of gene expression regulation[37]. Specific tiRNAs, derived from tRNA-Ala and tRNA-Cys, have been shown to inhibit protein translation by assembling into a G-quadruplex-like structure (G4-motif) and promoting stress granules formation[38][39][40][41].
2.3. Long Non-Coding RNAs
The ENCODE project established the existence of 9000–50,000 lncRNAs in the human genome. Transcribed from intergenic regions, from gene regulatory regions and from specific chromosomal regions by RNA polmymerase II, they are usually subjected to capping and alternative splicing [7][42]. lncRNAs’ main functions encompass the control of gene expression at different levels—both transcriptional and translational—the control of chromatin folding and the recruitment of chromatin modifiers[43][44]. Nevertheless, although several lncRNAs have been well characterized to date, further research is still required to understand their myriad of functions.
Forty percent of described lncRNAs have been reported in the CNS[45], where they participate in gene expression, imprinting and pluripotency regulation, as reported by numerous studies [46][47][48][49]. One of the first described lncRNAs was H19, a paternally imprinted gene, whose importance in embryonic development and neural viability has been established[50][51]. Other lncRNAs, such as Evf2 and MALAT1, are highly abundant in neurons showing a dynamic expression pattern in cell differentiation[47][52]. NeuroLNC, a nuclear lncRNA, has been recently involved in synapsis establishment[49]. In addition, RMST1 functions as a guide-RNA for SOX2 transcription factor towards neurogenic genes[53] promoting neural differentiation although it is also expressed in astrocytes [54]. Similarly, Pnky lncRNA, a highly conserved lncRNA, is very abundant in neural stem cells, regulating alternative splicing and cell fate decisions. Ramos and colleagues described it as essential for neural cell differentiation and renewal[55]. Another highly conserved lncRNA, Cyrano, is as well enriched in the nervous system, being a very important molecule for embryonic stem cells maintenance. Kleaveland et al. determined a whole functional circuit for Cyrano in which it represses miR-7 expression and avoids CDR1-AS destruction, a circular RNA known to regulate neuronal activity[59]. On the other hand, NEAT1, a lncRNA mostly expressed in oligodendrocytes and astrocytes, regulates alternative splicing and has been related to the neural response to stress.
2.4. Circular RNAs (circRNAs)
Circular RNAs (circRNAs), are the only type of ncRNA with covalently linked ends. Usually derived from protein-coding genes and single-stranded, they are produced by a non-canonical form of splicing, the so-called back-splicing[57][58]. This extraordinarily stable class of ncRNAs have been found in diverse species, from prokaryotes to eukaryotes, being expressed in a tissue-specific manner. circRNAs are generally found in the cytoplasm where they mostly act as miRNA sponges[59]. Other attributed circRNA functions comprise the regulation of riboproteins and RNA interaction, the transport of miRNAs inside the cells or the regulation of cell differentiation and proliferation [59].
Similar to other ncRNAs, circRNAs are over-represented in the nervous system, showing a differential expression pattern along neurodevelopmental stages and in the distinct brain regions. As recently reviewed somewhere else [60], the high expression of circRNAs during neurogenesis and nervous system development has been already reported[60][61] and their role in synaptic transmission has been widely explored[61][62][63].
3. NcRNAs and Oxidative Stress Management in Neurodegenerative Diseases
Diverse classes of ncRNAs are involved in OS signaling pathways. The general oxidation of RNAs, including the most abundant classes of ncRNAs (rRNAs and tRNAs) is an early event in major neurodegenerative diseases (reviewed in[64]). However, the understanding of the impact of oxidized rRNA on cell physiology is still very limited [65]. Other classes of housekeeping ncRNAs (snRNAs and snoRNAs) are also altered in response to ROS accumulation[66], with specific species modulating the cell response to OS in cancer[67]. Interestingly, defects in snoRNAs are associated with neurodevelopmental disorders such as Prader–Willi syndrome[68]. Moreover, several works have identified small RNA molecules generated through the processing of snoRNAs (sno-derived RNAs), specifically involved in gene silencing[69]. Nevertheless, although snRNAs and snoRNAs are altered in neurodegenerative conditions, their direct involvement in OS management and neuronal dysfunction needs further research efforts.
Compelling evidence exists involving the regulatory ncRNAs in OS-neurodegeneration pathways. Regulatory ncRNA expression is sensitive to ROS accumulation, with these perturbations contributing to neuronal dysfunction and microglial activation. miRNAs are the most intensively studied class, and different types are up- and downregulated in response to OS; however, analogous studies regarding other classes of ncRNAs are scarce. The dynamic changes in the concentration of specific ncRNA that target important genes for the control of ROS homeostasis and metabolism, together with the intrinsic differential susceptibility of cells to ROS-damage, underlie the cell capability to cope with sustained ROS production. This is particularly relevant in neuronal cells with increased susceptibility to OS as explained in the previous section.
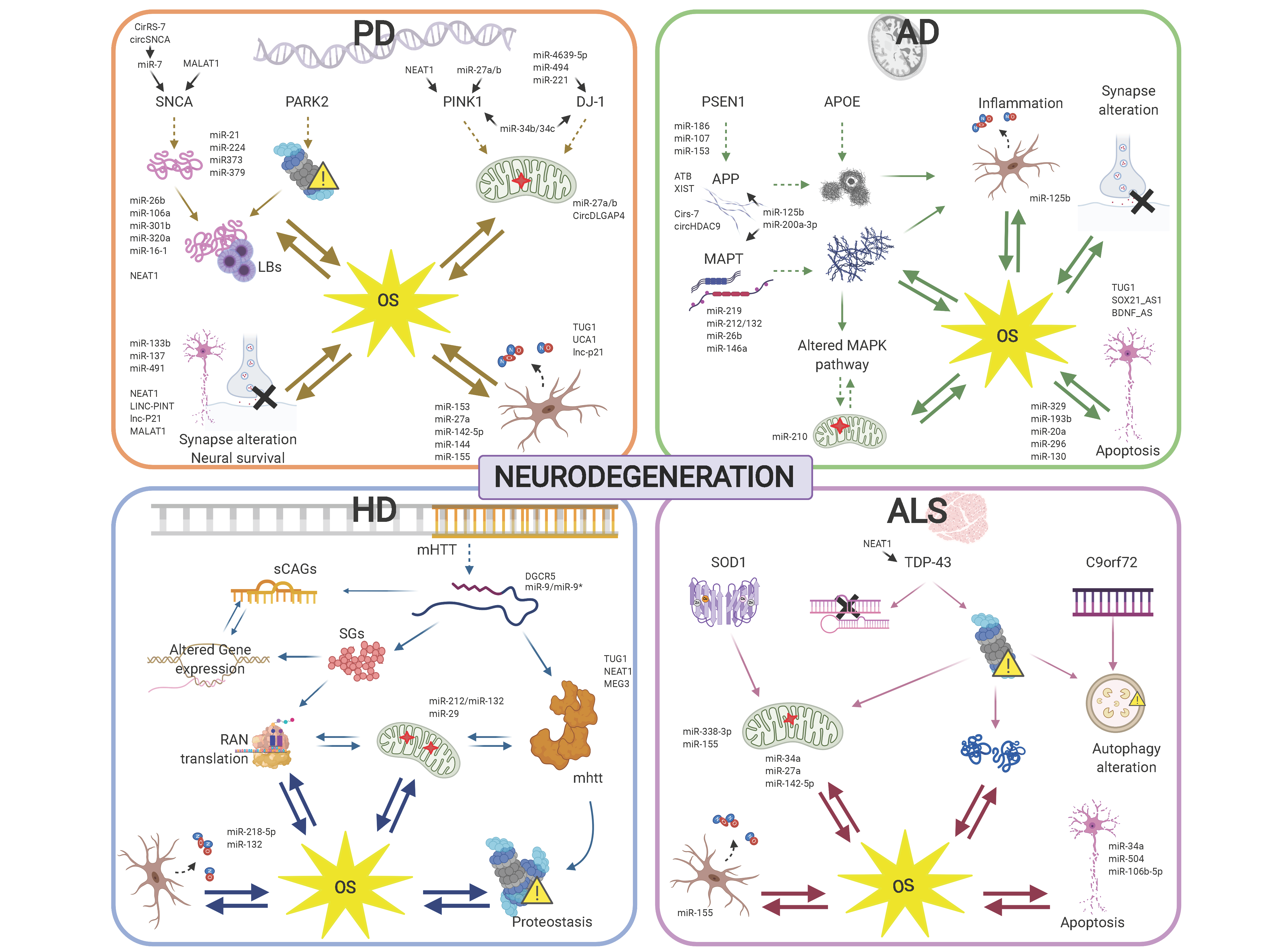
Specific ncRNA pathways regulating the cell response to OS are similarly altered in diverse neurodegenerative conditions, which may underlie commonly perturbed pathways, including mitochondrial dysfunction and the spread of insoluble protein inclusions. However, the altered activity of ncRNAs specifically targeting disease-related genes provides a scenario highlighting the differences in the development and progression of the different neurodegenerative diseases (Figure 2).
In this review, we have focused on regulatory ncRNA deregulation with direct implications in OS in the context of major neurodegenerative conditions: Alzheimer’s disease (AD), Parkinson’s disease (PD), Huntington’s disease (HD) and amyotrophic lateral sclerosis (ALS). Thus being the overall conclusions from the works presented here: (1) OS plays a major and early role in neurodegenerative diseases; (2) regulatory ncRNAs expression and activity dynamics regulate, directly and/or indirectly, gene expression networks involved in ROS homeostasis and buffering; (3) miRNAs are the best known class of ncRNAs, but other types of regulatory ncRNAs, including lncRNAs, tRFs, and circRNAs, appear as novel classes with a direct impact in OS modulation and neurodegeneration.
The entry is from 10.3390/antiox9111095
References
- Yaribeygi, H.; Panahi, Y.; Javadi, B.; Sahebkar, A. The underlying role of oxidative stress in neurodegeneration: A mechanistic review. CNS Neurol. Disord.-Drug Targets 2018, 17, 207–215.
- Feingold, E.A.; Good, P.J.; Guyer, M.S.; Kamholz, S.; Liefer, L.; Wetterstrand, K.; Collins, F.S.; Gingeras, T.R.; Kampa, D.; Sekinger, E.A.; et al. The ENCODE (ENCyclopedia of DNA Elements) project. Science 2004, 306, 636–640.
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Nie, L.; Wu, H.-J.; Hsu, J.-M.; Chang, S.-S.; Labaff, A.M.; Li, C.-W.; Wang, Y.; Hsu, J.L.; Hung, M.-C. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012, 4, 127–150.
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, biology and functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17.
- Salta, E.; De Strooper, B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012, 11, 189–200.
- Qureshi, I.A.; Mehler, M.F. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012, 13, 528–541.
- Wu, Y.-Y.; Kuo, H.-C. Functional roles and networks of non-coding RNAs in the pathogenesis of neurodegenerative diseases. J. Biomed. Sci. 2020, 27, 1–23.
- Ma, F.; Zhang, X.; Yin, K.-J. MicroRNAs in central nervous system diseases: A prospective role in regulating blood-brain barrier integrity. Exp. Neurol. 2020, 323, 113094.
- Duran, R.C.-D.; Wei, H.; Kim, D.H.; Wu, J.Q. Invited review: Long non-coding RNAs: Important regulators in the development, function and disorders of the central nervous system. Neuropathol. Appl. Neurobiol. 2019, 45, 538–556.
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874.
- Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002, 82, 47–95.
- Beckman, K.B.; Ames, B.N. The free radical theory of aging matures. Physiol. Rev. 1998, 78, 547–581.
- Bush, A.I. Metals and neuroscience. Curr. Opin. Chem. Biol. 2000, 4, 184–191.
- Jan, A.T.; Azam, M.; Siddiqui, K.; Ali, A.; Choi, I.; Haq, Q.M.R. Heavy metals and human health: Mechanistic insight into toxicity and counter defense system of antioxidants. Int. J. Mol. Sci. 2015, 16, 29592–29630.
- Calderón-Garcidueñas, L.; Leray, E.; Heydarpour, P.; Torres-Jardón, R.; Reis, J. Air pollution, a rising environmental risk factor for cognition, neuroinflammation and neurodegeneration: The clinical impact on children and beyond. Rev. Neurol. 2016, 172.
- Harwell, B. Biochemistry of oxidative stress. Biochem Soc Trans. 2007, 35 (Pt 5), 1147–1150.
- Echtay, K.S.; Roussel, D.; St-Pierre, J.; Jekabsons, M.B.; Cadenas, S.; Stuart, J.A.; Harper, J.A.; Roebuck, S.J.; Morrison, A.; Pickering, S.; et al. Superoxide activates mitochondrial uncoupling proteins. Nat. Cell Biol. 2002, 415, 96–99.
- Halliwell, B. Free radicals, antioxidants, and human disease: Curiosity, cause, or consequence? Lancet 1994, 344, 721–724.
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 1–23.
- Chmielarz, P.; Konovalova, J.; Najam, S.S.; Alter, H.; Piepponen, T.P.; Erfle, H.; Sonntag, K.C.; Schütz, G.; Vinnikov, I.A.; Domanskyi, A. Dicer and microRNAs protect adult dopamine neurons. Cell Death Dis. 2017, 8, e2813.
- Zhao, X.; He, X.; Han, X.; Yu, Y.; Ye, F.; Chen, Y.; Hoang, T.; Xu, X.; Mi, Q.-S.; Xin, M.; et al. MicroRNA-Mediated control of oligodendrocyte differentiation. Neuron 2010, 65, 612–626.
- Dugas, J.C.; Cuellar, T.L.; Scholze, A.; Ason, B.; Ibrahim, A.; Emery, B.; Zamanian, J.L.; Foo, L.C.; McManus, M.T.; Barres, B.A. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron 2010, 65, 597–611.
- Pogue, A.; Cui, J.; Li, Y.; Zhao, Y.; Culicchia, F.; Lukiw, W. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci. Lett. 2010, 476, 18–22.
- Taj, S.H.; Kho, W.; Riou, A.; Wiedermann, D.; Hoehn, M. MiRNA-124 induces neuroprotection and functional improvement after focal cerebral ischemia. Biomaterials 2016, 91, 151–165.
- Guo, Y.; Hong, W.; Wang, X.; Zhang, P.; Körner, H.; Tu, J.; Wei, W. MicroRNAs in microglia: How do MicroRNAs affect activation, inflammation, polarization of microglia and mediate the interaction between microglia and glioma? Front. Mol. Neurosci. 2019, 12, 125.
- Kumar, P.; Kuscu, C.; Dutta, A. biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016, 41, 679–689.
- Yamasaki, S.; Ivanov, P.; Hu, G.-F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42.
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. TRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 2015, 43, D141–D145.
- Guzzi, N.; Bellodi, C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol. 2020, 17, 1214–1222.
- Karaiskos, S.; Grigoriev, A. Dynamics of tRNA fragments and their targets in aging mammalian brain. F1000Research 2016, 5, 2758.
- Zhang, S.; Li, H.; Zheng, L.; Li, H.; Feng, C.; Zhang, W. Identification of functional tRNA-derived fragments in senescence-accelerated mouse prone 8 brain. Aging 2019, 11, 10485–10498.
- Krishna, S.; Yim, D.G.; Lakshmanan, V.; Tirumalai, V.; Koh, J.L.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.J.; Sze, S.K.; et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019, 20.
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J. Hematol. Oncol. 2020, 13, 1–14.
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409.
- Guan, L.; Karaiskos, S.; Grigoriev, A. Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA Biol. 2019, 17, 1070–1080.
- Kumar, P.; Anaya, J.; Mudunuri, S.B.; Dutta, A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014, 12, 78.
- Gonskikh, Y.; Gerstl, M.; Kos, M.; Borth, N.; Schosserer, M.; Grillari, J.; Polacek, N. Modulation of mammalian translation by a ribosome-associated tRNA half. RNA Biol. 2020, 17, 1125–1136.
- Wolozin, B.; Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666.
- Ivanov, P.; O’Day, E.; Emara, M.M.; Wagner, G.; Lieberman, J.; Anderson, P.J. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA 2014, 111, 18201–18206.
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623.
- Lyons, S.M.; Achorn, C.; Kedersha, N.L.; Anderson, P.J.; Ivanov, P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016, 44, 6949–6960.
- Salvatori, B.; Biscarini, S.; Morlando, M. Non-coding RNAs in nervous system development and disease. Front. Cell Dev. Biol. 2020, 8, 8.
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46.
- Briggs, J.A.; Wolvetang, E.J.; Mattick, J.S.; Rinn, J.L.; Barry, G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 2015, 88, 861–877.
- Pollard, K.S.; Salama, S.R.; Lambert, N.; Lambot, M.-A.; Coppens, S.; Pedersen, J.S.; Katzman, S.; King, B.; Onodera, C.; Siepel, A.; et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nat. Cell Biol. 2006, 443, 167–172.
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093.
- Hart, R.P.; Goff, L.A. Long noncoding RNAs: Central to nervous system development. Int. J. Dev. Neurosci. 2016, 55, 109–116.
- Keihani, S.; Kluever, V.; Mandad, S.; Bansal, V.; Rahman, R.; Fritsch, E.; Gomes, L.C.; Gärtner, A.; Kügler, S.; Urlaub, H.; et al. The long noncoding RNA neuroLNC regulates presynaptic activity by interacting with the neurodegeneration-associated protein TDP-43. Sci. Adv. 2019, 5, eaay2670.
- Yang, C.; Tang, R.; Ma, X.; Wang, Y.; Luo, D.; Xu, Z.; Zhu, Y.; Yang, L. Tag SNPs in long non-coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population. Oncotarget 2015, 6, 15311–15320.
- Han, C.-L.; Ge, M.; Liu, Y.-P.; Zhao, X.-M.; Wang, K.-L.; Chen, N.; Meng, W.-J.; Hu, W.; Zhang, J.-G.; Li, L.; et al. LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J. Neuroinflamm. 2018, 15, 1–9.
- Bond, A.M.; VanGompel, M.J.W.; Sametsky, E.A.; Clark, M.F.; Savage, J.C.; Disterhoft, J.F.; Kohtz, J.D. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 2009, 12, 1020–1027.
- Ng, S.-Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359.
- Reddy, A.S.; O’Brien, D.; Pisat, N.; Weichselbaum, C.T.; Sakers, K.; Lisci, M.; Dalal, J.S.; Dougherty, J.D. A comprehensive analysis of cell type–specific nuclear RNA from neurons and glia of the brain. Biol. Psychiatry 2017, 81, 252–264.
- Ramos, A.D.; Andersen, R.E.; Liu, S.J.; Nowakowski, T.J.; Hong, S.J.; Gertz, C.C.; Salinas, R.D.; Zarabi, H.; Kriegstein, A.R.; Lim, D.A. The long noncoding RNA pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015, 16, 439–447.
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A network of noncoding regulatory RNAs Acts in the mammalian brain. Cell 2018, 174, 350–362.e17.
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Yu, C.-Y.; Kuo, H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 1–12.
- Mehta, S.L.; Dempsey, R.J.; Vemuganti, R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020, 186, 101746.
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.J.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610.
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Jara, C.A.C.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357.
- Zhang, Y.; Du, L.; Bai, Y.; Han, B.; He, C.; Gong, L.; Huang, R.; Shen, L.; Chao, J.; Liu, P.; et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry 2018, 25, 1175–1190.
- Liu, Z.; Chen, X.; Li, Z.; Ye, W.; Ding, H.; Li, P.; Aung, L.H.H. Role of RNA oxidation in neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 5022.
- Shcherbik, N.; Pestov, D.G. The impact of oxidative stress on ribosomes: From injury to regulation. Cells 2019, 8, 1379.
- Leisegang, M.S.; Schröder, K.; Brandes, R.P. Redox regulation and noncoding RNAs. Antioxidants Redox Signal. 2018, 29, 793–812.
- Chu, L.; Su, M.Y.; Maggi, L.B., Jr.; Lu, L.; Mullins, C.; Crosby, S.; Huang, G.; Chng, W.J.; Vij, R.; Tomasson, M.H. Multiple myeloma–associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J. Clin. Investig. 2012, 122, 2793–2806.
- Sahoo, T.; Del Gaudio, D.; German, J.R.; Shinawi, M.; Peters, S.U.; Person, R.E.; Garnica, A.D.; Cheung, S.W.; Beaudet, A.L. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008, 40, 719–721.
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs derived from snoRNAs. RNA 2009, 15, 1233–1240.