Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Tingdong Li and Version 4 by Camila Xu.
Herpesviruses are major pathogens that infect humans and animals. Manipulating the large genome is critical for exploring the function of specific genes and studying the pathogenesis of herpesviruses and developing novel anti-viral vaccines and therapeutics. 疱疹病毒是感染人类和动物的主要病原体。操纵大基因组对于探索特定基因的功能和研究疱疹病毒的发病机制以及开发新型抗病毒疫苗和治疗方法至关重要。细菌人工染色体(Bacterial artificial chromosome (BAC) technology significantly advanced the capacity of herpesviruses researchers to manipulate the virus genomes.AC)技术显着提高了疱疹病毒研究人员操纵病毒基因组的能力。
- herpesvirus
- bacterial artificial chromosomes
- gene editing
- virus mutant selection
1. Introduction 简介
Genome manipulation is an effective method for studying the function of viral genes and can help scientists understand the biology of viruses, such as discovering virulence factors and exploring new targets for developing novel vaccines and antiviral drugs for the prevention and treatment of viral infection. Herpesviruses are a group of double-strand 基因组操作是研究病毒基因功能的有效方法,可以帮助科学家了解病毒的生物学,如发现毒力因子,探索开发预防和治疗病毒感染的新型疫苗和抗病毒药物的新靶点。疱疹病毒是一组双链DNA ((dsDNA) viruses which can cause lifelong persistent infections and are major pathogens in humans and a wide range of animals. The genomes of herpesviruses are between 125 and 295 kilobases in length [1], which makes it difficult for genome manipulation.
In the past decades, scientists have developed several methods for manipulating the genome of herpesviruses, such as )病毒,可引起终身持续感染,是人类和各种动物的主要病原体。疱疹病毒的基因组长度在125至295千碱基之间[1],这使得基因组操作变得困难。
在过去的几十年里,科学家们开发了几种操纵疱疹病毒基因组的方法,如λ-red, Cre-、Cre-loxp, 、CRISPR-Cas9, and others, which helped understand the pathogenesis of herpesviruses and develop novel vaccines and antiviral drugs [2][3]. Traditionally, linear 等,有助于了解疱疹病毒的发病机制,开发新型疫苗和抗病毒药物[2,3]。传统上,将含有选择盒和侧翼同源区域的线性DNA fragments or circular plasmids containing selection cassettes and flanking homologous regions were transferred into herpesvirus-infected cells, and the genomes of herpesviruses were edited by recombination, and then the recombinant viruses were selected by using different markers (different selection cassettes) [4][5][6][7]片段或环状质粒转移到疱疹病毒感染的细胞中,通过重组编辑疱疹病毒的基因组,然后使用不同的标记(不同的选择盒)选择重组病毒[4,5,6,7]. This approach has a low efficiency. In addition, its dependence on the production of progeny viruses results in a limited range of genes that can be edited. Moreover, the cumbersome screening process for identifying functional recombinant viruses restricts its application [3]. Therefore, it is imperative to explore alternative techniques that can enhance the efficiency and expand the range of genes that can be edited. 这种方法效率低。此外,它对后代病毒产生的依赖导致可以编辑的基因范围有限。此外,用于识别功能性重组病毒的繁琐筛选过程限制了其应用[3]。因此,必须探索能够提高效率和扩大可编辑基因范围的替代技术。基于大肠杆菌因子F构建的细菌人工染色体(Bacterial artificial chromosomes (BAC), constructed based on factor F of Escherichia coli (E. coli), provide an important technical platform for the editing of large genomes, including herpesviruses [8][9][10][11]. )为编辑包括疱疹病毒在内的大基因组提供了重要的技术平台[8,9,10,11]。基于BAC-based gene editing strategies are independent of the production of the progeny, and the screening efficiency is higher than conventional strategies [12][13][14]. Firstly, 的基因编辑策略独立于后代的产生,筛选效率高于常规策略[12,13,14]。首先,BACs can accommodate the insertion and stable inheritance of exogenous gene fragments ranging from 可以容纳100 to -300 kb. The F factor facilitates the maintenance of low copy numbers (外源基因片段的插入和稳定遗传。F因子有助于维持大肠杆菌中的低拷贝数(每个细胞1–-2 copies per cell) in E. coli which greatly reduces the difficulty in screening. Meanwhile, the development of various gene editing techniques in E. coli has greatly enhanced the editing efficiency of herpesvirus 个拷贝),从而大大降低了筛选的难度。同时,各种基因编辑技术在大肠杆菌中的开发,与传统方法相比,大大提高了疱疹病毒BACs compared to traditional methods. Edited 的编辑效率。编辑过的BACs can be reintroduced into eukaryotic cells to reconstruct herpesvirus mutants for further study of their biological behaviors in cells. In addition, the selection of recombinantly engineered viruses is not dependent on the generation of progeny viruses, allowing editing of the γ herpes virus genome that maintains latent infection in the long term [13][15] as well as manipulating genes essential for the production of progeny viruses [11].
可以重新引入真核细胞以重建疱疹病毒突变体,以进一步研究它们在细胞中的生物学行为。此外,重组工程病毒的选择不依赖于后代病毒的产生,允许编辑长期维持潜伏感染的γ疱疹病毒基因组[13,15]以及操纵产生后代病毒所必需的基因[11]。
2. Genome Editing Techniques of Herpesviruses Based on B基于BAC的疱疹病毒基因组编辑技术
2.1. RecA Recombination Technique
2.1. RecA 重组技术
The RecA recombination technique is a bacterial endogenous homologous recombination system composed of RecA recombinase and related auxiliary proteins (such as 重组技术是由RecA重组酶和相关辅助蛋白(如RecBCD, 、RecFOR, 、RuvABC, etc.). 等)组成的细菌内源同源重组系统。RecBCD is a multifunctional enzyme complex consisting of RecB, RecC, and RecD. When binding to the incision on the double-stranded donor DNA, RecB and RecD exhibit 3′ to 5′ and 5′ to 3′ helicase activities, respectively, which together result in the opening of the complementary double-stranded DNA (Figure 1是一种多功能酶复合物,由RecB,RecC和RecD组成。当与双链供体DNA上的切口结合时,RecB和RecD分别表现出3′至5′和5′至3′解旋酶活性,它们共同导致互补双链DNA的开放(图1a) [16]. Then, )[16]。然后,RecC can mediate the recognition of chi sites in the sequence and there可以介导序列中气位点的识别,从而引起变构改变,导致运动速度降低(图1by cause allosteric changes, leading to a decrease in the pace of movement (Figure 1b) [17]. At this point, )[17]。此时,RecB also has exonuclease activity, which mediates the generation of single-stranded 还具有核酸外切酶活性,其介导3′末端单链DNA at the 3′ end (Figure 的生成(图1c)[18,1c) [18][19][20]. The 9,20]。RecA recombinase binds to single-stranded 重组酶与核酸外切酶产生的单链DNA sites generated by the exonucleases and engages in the process of identifying homologous regions on the BAC for subsequent annealing and interaction. After successful interaction, the nucleoprotein filaments invade the dsDNA and undergo strand exchange to form heteroduplex DNA (Figure 位点结合,并参与鉴定BAC上同源区域的过程,以便随后退火和相互作用。成功相互作用后,核蛋白丝侵入dsDNA并进行链交换以形成异质双链DNA(图1d). Finally, )。最后,RuvABC assists in catalyzing branch migration and degradation of the heteroduplex DNA, resulting in homologous recombination (Figur有助于催化分支迁移和异源双链DNA的降解,导致同源重组(图1e 1e) [21].)[21]。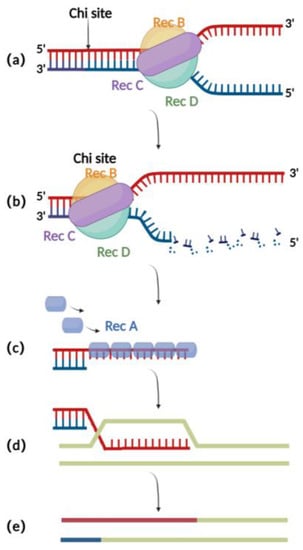
Figure 图1 . Schematic diagram of RecA recombination technique:重组技术示意图:(a)RecBCD binds to the double-stranded 与双链DNA terminus and promotes unwinding;末端结合并促进解开。(b)At the chi site, a single-stranded 在气位点,在3'末端产生单链DNA is produced at the 3′ end;。(c) The RecA protein binds to single-stranded DNA at the 3′ end;蛋白在3'末端与单链DNA结合。(d)ssDNA-RecA invades intact homologous double-stranded 侵入完整的同源双链DNA for strand exchange;进行链交换。(e) Recombination completed.重组完成。
Table表 1.The pros and cons of BAC-based herpesvirus genome editing techniques.
基于BAC的疱疹病毒基因组编辑技术的优缺点。
| Editing Techniques编辑技巧 | Length of Homologous Arms同源臂的长度 | Editing Efficiency编辑效率 | Requirement for Editing Site编辑网站的要求 | Stability of BAC的稳定性 | |
|---|---|---|---|---|---|
| RecA Recombination | RecA 重组 | 500 bp基点–3 k bp千基点 | 10−6 to到 10−4 | None没有 | Causes导致 mutations in the BAC 突变 |
| λ-red Recombination | λ-红色复合 | 30–50 bp基点 | <1% | None没有 | Maintained the stability of 保持了BAC的稳定性 |
| Base Editing基本编辑 | Not Required | 不需要 | Approaching接近 100% | PAM Sites & Base Editing Sites网站和基本编辑网站; Only Base Editing can be done只能进行基本编辑 | Maintained the stability of 保持了BAC的稳定性 |
2.2. λ-Red Recombination Technique
2.2. λ-红色复合技术
Homologous recombination mediated by the 由λ-red recombination system is currently the most widely used recombination technique for editing herpesvirus BAC clones [2][25]. The 重组系统介导的同源重组是目前编辑疱疹病毒BAC克隆最广泛使用的重组技术[2,25]。λ-red recombination system is derived from the phage 红色重组系统来源于噬菌体λ and consists of ,由Gam, Exo, and Beta proteins. The Gam protein is an auxiliary protein of Exo protein and Beta protein. The Gam protein can inhibit the binding of 、Exo和Beta蛋白组成。Gam蛋白是Exo蛋白和β蛋白的辅助蛋白。Gam蛋白可以抑制Rec BCD to the ends of dsDNA and thus inhibit the function of 与dsDNA末端的结合,从而抑制Rec BCD exonuclease, preventing the degradation of the exogenous double-stranded DNA (Figure 外切酶的功能,防止外源性双链DNA的降解(图2a)[26,2a) [26][27]. 7]。Exo proteins can bin蛋白可以结合到d to the ends of a dsDNA donor, which contains homologous fragments on both sides of the target gene. Concurrently, Exo protein possesses 5′ to 3′ exonuclease activity, producing a stretch of single-stranded DNA at the 3′ end (Figure 2sDNA供体的末端,dsDNA供体在靶基因的两侧都包含同源片段。同时,Exo蛋白具有5′至3′核酸外切酶活性,在3′末端产生一段单链DNA(图2b) [28]. Beta protein plays a decisive role in the process of )[28]。β蛋白在λ-red homologous recombination. As a single-stranded 红同源重组过程中起决定性作用。作为单链DNA binding protein, the beta protein binds to the single-stranded DNA produced by the Exo protein. The binding of Beta protein enhances the annealing of the donor DNA fragment and the homologous sequence at the target site of the replicating herpes virus BAC (Figure 结合蛋白,β蛋白与Exo蛋白产生的单链DNA结合。β蛋白的结合增强了供体DNA片段的退火和复制疱疹病毒BAC靶位点的同源序列(图2c). Homologous recombination is complete with the replication of )。DNA (Figure 2的复制完成了同源重组(图2d) [29][30].)[29,30]。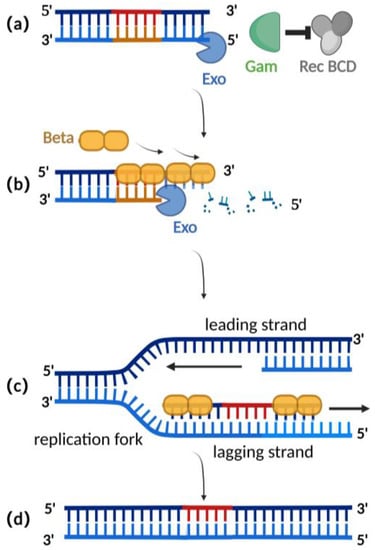
Figure 2. Schematic diagram of λ-red recombination technique: (a) Gam protein inhibits the activity of Rec BCD exonuclease; (b) Exo protein creates a stretch of single-stranded DNA at the 3′ end; (c) Beta proteins bind here, facilitating annealing interactions between the donor DNA fragment and the homologous sequence of the target site; (d) homologous recombination is complete with the replication of DNA.
2.3. Base Editing Technique
CRISPR/Cas 9 is a powerful genome editing technique [36] that has been successfully applied in a wide range of eukaryotic cells, including human cell lines, embryonic stem cells, mice, Arabidopsis, and Drosophila [37][38][39][40][37,38,39,40]. In 2013, Jiang et al. successfully edited Streptococcus pneumonia’s genome using CRISPR/Cas-9-only gene editing [41]. Since then, it has been successfully employed in a variety of prokaryotic species as well [42][43][42,43]. The versatility and effectiveness of CRISPR/Cas9 in modifying the genome make it a valuable tool for a wide range of scientific and medical applications. Due to the lack of the non-homologous end-joining (NHEJ) pathway [44][45][46][47][44,45,46,47] and the low efficiency of their endogenous homologous recombination system, it is difficult to achieve stable genome editing in most bacteria using CRISPR/Cas9 gene editing technology alone [41][48][49][50][41,48,49,50]. Until now, there was still no publication reporting CRISPR/Cas 9-based gene editing technique in the stably preserved herpesvirus BAC gene in E. coli. This highlights the need to explore alternative approaches to achieve efficient and reliable gene editing in these organisms. Recently, scientists have constructed an efficient gene editing method by combing CRISPR/Cas 9 gene editing technique with precise base editing technology. This method consists of an sgRNA and a complex that includes modified Cas9 proteins, cytosine deaminases, and an uracil glycosylase inhibitor (UGI) [51]. Unlike wild-type Cas9 proteins, the modified Cas9 is catalytically dead, lacking endonuclease activity. Therefore, it can only facilitate genome targeting via sgRNA but cannot induce a double strand break (DSB) due to the absence of cleavage activity (Figure 3a). Cytosine deaminase converts the specified cytosine (C) site to uracil (U) (Figure 3b). At this point, uracil glycosylase inhibitors can prevent the excision of intermediate product U, increasing the efficiency of converting C to T on the DNA chain, ultimately achieving single-base precise editing of C to T and G to A (Figure 3c).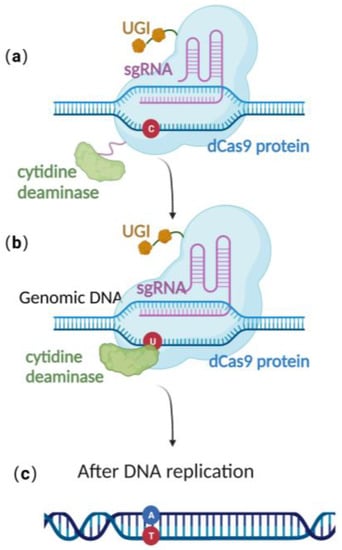
Figure 3. Schematic diagram of base editing method: (a) dCas9 mediates targeting without DSB formation; (b) cytosine deaminase converts C to U; (c) single-base precise editing is complete with the replication of DNA.
