The SARS-CoV-2 (severe acute respiratory syndrome coronavirus responsible for the COVID-19 disease) uses the Spike proteins of its envelope for infecting target cells expressing on the membrane the angiotensin converting enzyme 2 (ACE2) enzyme that acts as a receptor. To control the pandemic, genetically engineered vaccines have been designed for inducing neutralizing antibodies against the Spike proteins. These vaccines do not act like traditional protein-based vaccines, as they deliver the message in the form of mRNA or DNA to host cells that then produce and expose the Spike protein on the membrane (from which it can be shed in soluble form) to alert the immune system. Since the genetically engineered vaccines have been modified to be more stable and because they are distributed via liponanoparticles to various tissues and organs, this type of products may cause unforeseen serious adverse effects.
- Spike
- vaccine
- immune response
- thrombosis
- myocarditis
- inflammation
- renin-angiotensin system
- COVID-19
- SARS-CoV-2
1. Essentials of mRNA Vaccines Design and Functioning
2. The Immune Response to the SARS-CoV-2 and to the mRNA Vaccines
2.1. The Importance of the Route of Entry
2.2. Immunization Pathways of the SARS-CoV-2 and mRNA Vaccines
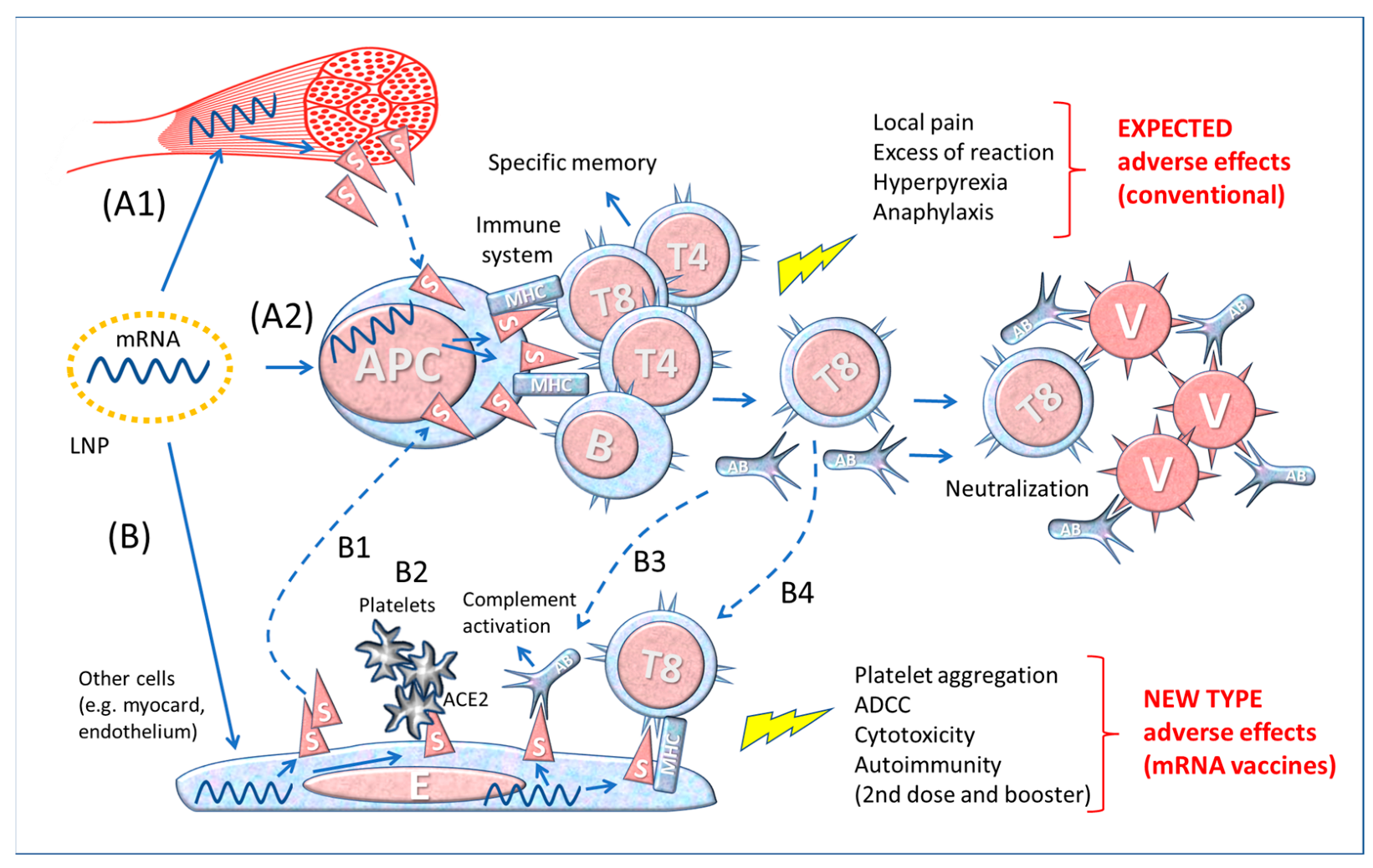
2.3. Differences between Contact with the Whole Virus and Vaccine-Derived Spike Protein
References
- Evans, J.P.; Liu, S.L. Role of host factors in SARS-CoV-2 entry. J. Biol. Chem. 2021, 297, 100847.
- Campbell, R.A.; Boilard, E.; Rondina, M.T. Is there a role for the ACE2 receptor in SARS-CoV-2 interactions with platelets? J. Thromb. Haemost. 2021, 19, 46–50.
- Bugatti, A.; Filippini, F.; Bardelli, M.; Zani, A.; Chiodelli, P.; Messali, S.; Caruso, A.; Caccuri, F. SARS-CoV-2 Infects Human ACE2-Negative Endothelial Cells through an alphavbeta3 Integrin-Mediated Endocytosis Even in the Presence of Vaccine-Elicited Neutralizing Antibodies. Viruses 2022, 14, 705.
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279.
- Malone, R.W.; Felgner, P.L.; Verma, I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA 1989, 86, 6077–6081.
- Seneff, S.; Nigh, G.; Kyriakopoulos, A.M.; McCullough, P.A. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem. Toxicol. 2022.
- Forchette, L.; Sebastian, W.; Liu, T. A Comprehensive Review of COVID-19 Virology, Vaccines, Variants, and Therapeutics. Curr. Med. Sci. 2021, 41, 1037–1051.
- Heinz, F.X.; Stiasny, K. Distinguishing features of current COVID-19 vaccines: Knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines 2021, 6, 1–13.
- Trougakos, I.P.; Terpos, E.; Alexopoulos, H.; Politou, M.; Paraskevis, D.; Scorilas, A.; Kastritis, E.; Andreakos, E.; Dimopoulos, M.A. Adverse effects of COVID-19 mRNA vaccines: The spike hypothesis. Trends Mol. Med. 2022, 28, 542–554.
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094.
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175.
- De Beuckelaer, A.; Pollard, C.; Van Lint, S.; Roose, K.; Van Hoecke, L.; Naessens, T.; Udhayakumar, V.K.; Smet, M.; Sanders, N.; Lienenklaus, S.; et al. Type I Interferons Interfere with the Capacity of mRNA Lipoplex Vaccines to Elicit Cytolytic T Cell Responses. Mol. Ther. 2016, 24, 2012–2020.
- Andries, O.; Mc Cafferty, S.; De Smedt, S.C.; Weiss, R.; Sanders, N.N.; Kitada, T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control Release 2015, 217, 337–344.
- Park, J.W.; Lagniton, P.N.; Liu, Y.; Xu, R.-H. mRNA vaccines for COVID-19: What, why and how. Int. J. Biol. Sci. 2021, 17, 1446–1460.
- Kyriakopoulos, A.M.; McCullough, P.A. Synthetic mRNAs; Their Analogue Caps and Contribution to Disease. Diseases 2021, 9, 57.
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T.; et al. Improving mRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3’ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27, 824–836.
- McKernan, K.; Kyriakopoulos, A.M.; McCullough, P.A. Differences in Vaccine and SARS-CoV-2 Replication Derived mRNA: Implications for Cell Biology and Future Disease. OSF Preprints 2021.
- Mauro, V.P.; Chappell, S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014, 20, 604–613.
- Ogata, A.F.; Cheng, C.-A.; Desjardins, M.; Senussi, Y.; Sherman, A.C.; Powell, M.; Novack, L.; Von, S.; Li, X.; Baden, L.R.; et al. Circulating Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vaccine Antigen Detected in the Plasma of mRNA-1273 Vaccine Recipients. Clin. Infect. Dis. 2022, 74, 715–718.
- Roltgen, K.; Nielsen, S.C.A.; Silva, O.; Younes, S.F.; Zaslavsky, M.; Costales, C.; Yang, F.; Wirz, O.F.; Solis, D.; Hoh, R.A.; et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 2022, 185, 1025–1040.e14.
- Fertig, T.E.; Chitoiu, L.; Marta, D.S.; Ionescu, V.-S.; Cismasiu, V.B.; Radu, E.; Angheluta, G.; Dobre, M.; Serbanescu, A.; Hinescu, M.E.; et al. Vaccine mRNA Can Be Detected in Blood at 15 Days Post-Vaccination. Biomedicines 2022, 10, 1538.
- Castruita, J.A.S.; Vest Schneider, U.; Mollerup, S.; Leineweber, T.D.; Weis, N.; Bukh, J.; Pedersen, M.S.; Westh, H. SARS-CoV -2 spike mRNA vaccine sequences circulate in blood up to 28 days after COVID -19 vaccination. APMIS 2023. Epub ahead of print.
- Shrestha, N.K.; Burke, P.C.; Nowacki, A.S.; Simon, J.F.; Hagen, A.; Gordon, S.M. Effectiveness of the Coronavirus Disease 2019 (COVID-19) Bivalent Vaccine. medRxiv 2022.
- Wagenhäuser, I.; Reusch, J.; Gabel, A.; Krone, L.B.; Kurzai, O.; Petri, N.; Krone, M. Bivalent BNT162b2mRNA original/Omicron BA.4-5 booster vaccination: Adverse reactions and inability to work compared to the monovalent COVID-19 booster. medRxiv 2022.
- Hwang, I. Cell-cell communication via extracellular membrane vesicles and its role in the immune response. Mol. Cells 2013, 36, 105–111.
- Bansal, S.; Perincheri, S.; Fleming, T.; Poulson, C.; Tiffany, B.; Bremner, R.M.; Mohanakumar, T. Cutting Edge: Circulating Exosomes with COVID Spike Protein Are Induced by BNT162b2 (Pfizer-BioNTech) Vaccination prior to Development of Antibodies: A Novel Mechanism for Immune Activation by mRNA Vaccines. J. Immunol. 2021, 207, 2405–2410.
- Hu, W.; Pasare, C. Location, location, location: Tissue-specific regulation of immune responses. J. Leukoc. Biol. 2013, 94, 409–421.
- Horwitz, D.A.; Zheng, S.G.; Gray, J.D. Natural and TGF-beta-induced Foxp3(+)CD4(+) CD25(+) regulatory T cells are not mirror images of each other. Trends Immunol. 2008, 29, 429–435.
- Sterlin, D.; Mathian, A.; Miyara, M.; Mohr, A.; Anna, F.; Claer, L.; Quentric, P.; Fadlallah, J.; Devilliers, H.; Ghillani, P.; et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021, 13, eabd2223.
- Drummer, H.E.; Van, H.; Klock, E.; Zheng, S.; Wei, Z.; Boo, I.; Center, R.J.; Li, F.; Bhat, P.; Ffrench, R.; et al. Dimeric IgA is a specific biomarker of recent SARS-CoV-2 infection. medRxiv 2021.
- Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Viant, C.; Gaebler, C.; Cipolla, M.; Hoffmann, H.H.; Oliveira, T.Y.; Oren, D.A.; et al. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021, 13, eabf1555.
- Sheikh-Mohamed, S.; Isho, B.; Chao, G.Y.C.; Zuo, M.; Cohen, C.; Lustig, Y.; Nahass, G.R.; Salomon-Shulman, R.E.; Blacker, G.; Fazel-Zarandi, M.; et al. Systemic and mucosal IgA responses are variably induced in response to SARS-CoV-2 mRNA vaccination and are associated with protection against subsequent infection. Mucosal. Immunol. 2022, 15, 799–808.
- Azzi, L.; Dalla Gasperina, D.; Veronesi, G.; Shallak, M.; Ietto, G.; Iovino, D.; Baj, A.; Gianfagna, F.; Maurino, V.; Focosi, D.; et al. Mucosal immune response in BNT162b2 COVID-19 vaccine recipients. eBioMedicine 2021, 75, 103788.
- Azzi, L.; Gasperina, D.D.; Veronesi, G.; Shallak, M.; Maurino, V.; Baj, A.; Gianfagna, F.; Cavallo, P.; Dentali, F.; Tettamanti, L.; et al. Mucosal immune response after the booster dose of the BNT162b2 COVID-19 vaccine. Ebiomedicine 2023, 88, 104435.
- Singanayagam, A.; Hakki, S.; Dunning, J.; Madon, K.J.; Crone, M.A.; Koycheva, A.; Derqui-Fernandez, N.; Barnett, J.L.; Whitfield, M.G.; Varro, R.; et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: A prospective, longitudinal, cohort study. Lancet Infect. Dis. 2022, 22, 183–195.
- Ivanova, E.N.; Devlin, J.C.; Buus, T.B.; Koide, A.; Shwetar, J.; Cornelius, A.; Samanovic, M.I.; Herrera, A.; Mimitou, E.P.; Zhang, C.; et al. SARS-CoV-2 mRNA vaccine elicits a potent adaptive immune response in the absence of IFN-mediated inflammation observed in COVID-19. medRxiv 2021.
- Committee, F.A. Vaccines and Related Biological Products Advisory Committee December 17, 2020 in, Food and Drug Administration. 2020. Available online: https://www.fda.gov/advisory-committees/advisory-committee-calendar/vaccines-and-related-biological-products-advisory-committee-december-17-2020-meeting-announcement (accessed on 24 July 2022).
- Moore, M.J. mRNA Platform and Mechanism of Action of mRNA-1273 in, FDA document: Emergency Use Authorization (EUA) Application for mRNA-1273. 2020. Available online: https://www.fda.gov/media/144583/download (accessed on 24 July 2022).
- Plotkin, S.A. Vaccines: The fourth century. Clin. Vaccine Immunol. 2009, 16, 1709–1719.
- Bellavite, P. Causality assessment of adverse events following immunization: The problem of multifactorial pathology. F1000Research 2020, 9, 170.
- Reif, D.M.; McKinney, B.A.; Motsinger, A.A.; Chanock, S.J.; Edwards, K.M.; Rock, M.T.; Moore, J.H.; Crowe, J.J.E. Genetic Basis for Adverse Events after Smallpox Vaccination. J. Infect. Dis. 2008, 198, 16–22.
- Poland, G.A.; Kennedy, R.B.; McKinney, B.A.; Ovsyannikova, I.G.; Lambert, N.D.; Jacobson, R.M.; Oberg, A.L. Vaccinomics, adversomics, and the immune response network theory: Individualized vaccinology in the 21st century. Semin. Immunol. 2013, 25, 89–103.
- Lin, Y.; He, Y. The ontology of genetic susceptibility factors (OGSF) and its application in modeling genetic susceptibility to vaccine adverse events. J. Biomed. Semant. 2014, 5, 19.
- Klein, N.P.; Lewis, E.; McDonald, J.; Fireman, B.; Naleway, A.; Glanz, J.; Jackson, L.A.; Donahue, J.G.; Jacobsen, S.J.; Weintraub, E.; et al. Risk factors and familial clustering for fever 7–10 days after the first dose of measles vaccines. Vaccine 2017, 35, 1615–1621.
- Asandei, A.; Mereuta, L.; Schiopu, I.; Park, J.; Seo, C.H.; Park, Y.; Luchian, T. Non-Receptor-Mediated Lipid Membrane Permeabilization by the SARS-CoV-2 Spike Protein S1 Subunit. ACS Appl. Mater. Interfaces 2020, 12, 55649–55658.
- Yonker, L.M.; Swank, Z.; Bartsch, Y.C.; Burns, M.D.; Kane, A.; Boribong, B.P.; Davis, J.P.; Loiselle, M.; Novak, T.; Senussi, Y.; et al. Circulating Spike Protein Detected in Post–COVID-19 mRNA Vaccine Myocarditis. Circulation 2023. Epub ahead of print.
- Verma, A.K.; Lavine, K.J.; Lin, C.-Y. Myocarditis after Covid-19 mRNA Vaccination. N. Engl. J. Med. 2021, 385, 1332–1334.
- Krug, A.; Stevenson, J.; Høeg, T.B. BNT162b2 Vaccine-Associated Myo/Pericarditis in Adolescents: A Stratified Risk-Benefit Analysis. Eur. J. Clin. Investig. 2022, 52, e13759.
- Karlstad, Ø.; Hovi, P.; Husby, A.; Härkänen, T.; Selmer, R.M.; Pihlström, N.; Hansen, J.V.; Nohynek, H.; Gunnes, N.; Sundström, A.; et al. SARS-CoV-2 Vaccination and Myocarditis in a Nordic Cohort Study of 23 Million Residents. JAMA Cardiol. 2022, 7, 600.
- Atalis, A.; Keenum, M.C.; Pandey, B.; Beach, A.; Pradhan, P.; Vantucci, C. Nanoparticle-delivered TLR4 and RIG-I agonists enhance immune response to SARS-CoV-2 subunit vaccine. J. Control Release 2022, 347, 476–488.
