Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Dean Liu and Version 1 by Annamaria Pratelli.
Canine coronavirus (CCoV) is a positive-strand RNA virus generally responsible for mild-to-severe gastroenteritis in dogs. In recent years, new CCoVs with acquired pathogenic characteristics have emerged, turning the spotlight on the evolutionary potential of CCoVs.
- coronavirus
- dog
- human
- recombination
1. Introduction
Canine coronavirus (CCoV) is an enveloped virus within the Coronaviridae family with a single-strand positive-sense RNA genome, approximately 27–32kb in size. Based on phylogenetic analyses and genomic structures, CoVs are currently classified within family four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, which is the largest known RNA genome, and Deltacoronavirus, which recognises bats, birds, and likely rodents as natural reservoirs [1]. CCoV belongs to the genus Alphacoronavirus, which also includes the prototype viruses: feline coronaviruses (FCoVs), transmissible gastroenteritis virus (TGEV), mink coronavirus (MinkCoV), ferret coronavirus (FRCoV), and alpaca respiratory coronavirus [2]. Two thirds of the CCoV genome from the 5′ end consists of two large overlapping open reading frames (ORFs), ORF1a and ORF1b, encoding the replicase protein, the viral RNA-dependent RNA polymerase, and viral proteases. The remaining genome from the 3′ end contains ORFs encoding the four major structural proteins, spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, and for the non-structural proteins ORF3a, ORF3b, ORF3c, ORF7a, and ORF7b [3]. The spike protein, which assembles into trimers on the virion surface to form the distinctive “crown” appearance, is the main inducer of neutralizing antibodies and plays an important role during viral entry, mediating the viral attachment to specific host cell receptors and the fusion between the envelope and the plasma membrane [4]. The S protein ectodomain shares the same organization in all CoVs and is organized in two distinct domains: S1, the N-terminal domain (NTD) responsible for receptor binding, and S2, the C-terminal domain (CTD) responsible for fusion. A notable difference between the S proteins of different CoVs is whether they are cleaved during viral replication. Most Alphacoronaviruses and Betacoronaviruses, with some exceptions, share an uncleaved protein, but the S protein of Gammacoronaviruses and some Betacoronaviruses is cleaved into S1 and S2 domains by a Golgi-resident protease [5]. The most represented and abundant viral structural protein is the M protein, a type III glycoprotein consisting of a short amino-terminal ectodomain, a triple-spanning transmembrane domain, and a long carboxyl-terminal inner domain [6]. It contributes to core stability and can play a role in neutralizing viral infectivity in the presence of complement [7]. The small E protein was recognised as a structural component of the CoVs and is thought to be important for viral envelope assembly [8]. The N protein is a basic phosphoprotein that forms the helical nucleocapsid that binds to the viral RNA and modulates RNA synthesis [4]. The additional ORFs encoding non-structural proteins vary among different CoVs in number, nucleotide sequence, and gene order [9].
2. Canine Coronavirus: Light and Shadow
CCoV is considered one of the main pathogens responsible for enteritis in dogs, wolves, foxes, and other canine species [11][10], but it can also affect other animals thanks to its ability to undergo recombination that ensures the proliferation of new strains with selective advantages over the parental genomes and its ability to easily cross interspecies barriers. This host species extension, as well as the variation in the cell tropism and the pathogenicity of the virus, is mainly related to the variability of the S protein, which is responsible for the emergence of new virus strains, serotypes, and subtypes [12][11]. Starting from its first description in 1971 during an epizootic infection in a canine military unit in Germany [13][12], CCoV rapidly spread worldwide and today appears to be enzootic with a variable prevalence ranging from 6.25% to 72.5% [14,15][13][14]. Although in the past CCoV has been overlooked and vaccination is still not recommended due to the absence of an effective challenge model, this virus is involved in the onset of moderate-to-severe enteritis. Dogs of all breeds and ages are susceptible to infection and, although the virus alone can be responsible for epizootics mainly in kennels and animal shelters, fatal infections are unusual unless mixed infections and/or overcrowding/unsanitary conditions exist [3]. Nevertheless, in the last few decades, CCoV has undergone mutations/recombination over time, changing tissue tropism and virulence and generating new genetically divergent strains, some of which possess pathogenic potential [16][15]. After highlighting point mutations affecting the M gene and multiple regions of the viral genome including ORF1a, ORF1b, and ORF5 in several CCoVs detected in pups with diarrhoea in Italy, the phylogenetic analysis on the inferred aa sequence of a region encompassing about 80% of the S gene of one of these strains (strain Elmo/02) clearly showed that strain Elmo/02 segregates with FCoVs of type I (sharing about 81% identity) rather than reference CCoVs and FCoVs of type II (sharing about 54% identity) [17,18,19][16][17][18]. Other features differentiate Elmo/02 from classical strains: (i) a potential cleavage site in the S protein shared with Betacoronavirus and Deltacoronavirus; (ii) an additional ORF (referred as ORF3) 624 nt in length and encoding a putative 207aa-protein—likely secreted from the infected cells as no transmembrane region was found—that is completely absent in FCoV type I and of which only remnants remain in the genomes of CCoV type II and TGEV [20][19]. Based on these observations, two genotypes of CCoV designated as CCoV types I and II were proposed as observed from the genomic analysis of the genes encoding the main structural proteins of the virus [18,21,22][17][20][21]. Elmo/02 was designated as the prototype of the newly recognized CCoV type I, and the reference strains (i.e., Insavc-1, K378) as CCoV type II [18][17]. Most detected CCoVs are type II viruses that are also easily cultivated in canine and feline cell lines. On the contrary, CCoV type I, to date, has been only detected by reverse transcription-polymerase chain reaction (RT-PCR) and quantitative PCR (q-PCR) [16][15]. Reports on the emergence of new CCoV strains have always occurred and have been constant over time. Wesley [23][22] demonstrated that the N-terminus of the S gene of strain UCD-1 is more closely related to TGEV than to CCoV, and Naylor et al. [24][23] identified a divergent CCoV strain, UWSMN-1, in Australia, characterized by the presence of gradually accumulating point mutations randomly interspersed throughout its genome. In 2002, a new CCoV, BGF10, with a highly divergent region at the amino-terminal domain of the M protein and a long non-structural protein 3b—250 aa long and associated with virulence in other CoVs—was isolated in the United Kingdom [25][24]. Possible recombination events also occurred in certain CCoV strains detected in a fatal outbreak of gastroenteritis in Swedish dogs. These virulent strains displayed the 5′ end and 3′ end of an S gene closely related to CCoV type I and CCoV type II, respectively, suggesting a possible recombination event between CCoV genotypes [26][25]. The most striking example of a hypervirulent strain was observed in 2005 in Italy and later in other countries. The detected strain, CB/05, designated pantropic CCoV type II, was associated with the onset of severe and haemorrhagic gastroenteritis, which was sometimes fatal [27,28,29,30][26][27][28][29]. Interestingly, the CB/05 strain—detected in other organs including the brain in addition to the intestines—has a degree of aa identity to CCoV type II in the 3′ end of the genome, although the S gene displayed the highest identity to FCoV type II, strain 79-1683. In addition, a 38-nt deletion in ORF3b was identified as a genetic marker of this pantropic CCoV type II strain, but no obvious genetic signatures responsible for the switch in pathogenicity were found [31][30]. A genetic analysis of the accessory gene ORF3 highlighted that TGEV originated from CCoV through cross-species transmission, pointing out that the evolution of TGEV, FCoVs, and CCoVs are closely related to each other. In fact, not only did CCoV give rise to FCoV type II through double homologous recombination events between the S and M genes [32][31], and recombination events with an ancestral CoV might have given rise to the appearance of FCoV type I and CCoV type I [19][18], but a further recombination between CCoV type II and TGEV has occurred in the very 5′ end of the S gene, generating a back recombination event with the appearance of a TGEV-like CCoV (Figure 1). Phylogenetic analysis showed that TGEV-like CCoVs formed a monophyletic group with the same cluster of TGEV and porcine respiratory CoV in the N terminus of the S gene; however, in the C terminus, they clustered together with CCoV-II isolates and separately from porcine respiratory and enteric CoVs. The recombinant TGEV-like virus was detected in the internal organs and in the intestinal contents of infected pups which died with acute gastroenteritis, underlining the pathogenic role of this virus [33][32]. In addition, during 2001–2008, the distribution of the TGEV-like CCoVs was assessed in the canine populations of different geographic areas of Europe, and approximately 20% of positive CCoV samples were characterized as TGEV-like CCoV, with a detection rate which varied according to geographic origin. The confirmation of TGEV-like CCoV circulation among dog populations has epidemiological implications for prophylaxis programs in dogs based on the administration of CCoV vaccines prepared with the classical CCoV type II strains, which may not provide protection against TGEV-like CCoV infection and should draw attention to the pathogenetic and epidemiological role of these recombinant CCoV viruses [34][33]. The identification of a TGEV-like CCoV characterized by a potential double recombination through partial S-gene exchange with TGEV induced the proposal of a new classification for CCoV type II, which is now further divided into two subtypes: CCoV type IIa (including classical enteric CCoVs) and CCoV type IIb (including TGEV-like CCoVs) [33][32].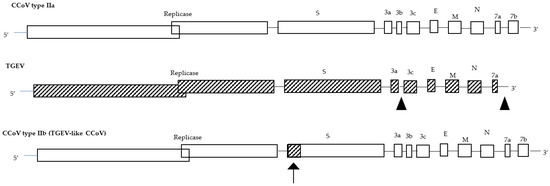
Figure 1. Genome organization of CCoV type IIa, TGEV, and CCoV type IIb (TGEV-like CCoV). The arrow shows the recombination site.
References
- Decaro, N.; Martella, V.; Saif, L.J.; Buonavoglia, C. COVID-19 from Veterinary Medicine and One Health Perspectives: What Animal Coronaviruses Have Taught Us. Res. Vet. Sci. 2020, 131, 21–23.
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Bamford, D.H.; Breitbart, M.; Davison, A.J.; Ghabrial, S.A.; Gorbalenya, A.E.; Knowles, N.J.; Krell, P.; et al. Ratification Vote on Taxonomic Proposals to the International Committee on Taxonomy of Viruses. Arch. Virol. 2015, 160, 1837–1850.
- Pratelli, A. The Evolutionary Processes of Canine Coronaviruses. Adv. Virol. 2011, 2011, 562831.
- Enjuanes, L.; Brian, D.; Cavanagh, D.; Holmes, K.; Lai, M.M.C.; Laude, H.; Masters, P.; Rottier, P.; Siddell, S.; Spaan, W.J.M.; et al. Virus Taxonomy; Murphy, F.A., Fauquet, C.M., Bishop, D.H.L., Ghabrial, S.A., Jarvis, A.W., Martelli, G.P., Mayo, M.A., Summers, M.D., et al., Eds.; Springer: Vienna, Austria, 1995; ISBN 978-3-211-82594-5.
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033.
- Rottier, P.J.M. The Coronavirus Membrane Glycoprotein. In The Coronaviridae; Springer: Boston, MA, USA, 1995; pp. 115–139.
- Escors, D.; Ortego, J.; Laude, H.; Enjuanes, L. The Membrane M Protein Carboxy Terminus Binds to Transmissible Gastroenteritis Coronavirus Core and Contributes to Core Stability. J. Virol. 2001, 75, 1312–1324.
- Vennema, H.; Godeke, G.J.; Rossen, J.W.; Voorhout, W.F.; Horzinek, M.C.; Opstelten, D.J.; Rottier, P.J. Nucleocapsid-Independent Assembly of Coronavirus-like Particles by Co-Expression of Viral Envelope Protein Genes. EMBO J. 1996, 15, 2020–2028.
- Lee, H.-J.; Shieh, C.-K.; Gorbalenya, A.E.; Koonin, E.v.; la Monica, N.; Tuler, J.; Bagdzhadzhyan, A.; Lai, M.M.C. The Complete Sequence (22 Kilobases) of Murine Coronavirus Gene 1 Encoding the Putative Proteases and RNA Polymerase. Virology 1991, 180, 567–582.
- Dong, B.; Zhang, X.; Bai, J.; Zhang, G.; Li, C.; Lin, W. Epidemiological Investigation of Canine Coronavirus Infection in Chinese Domestic Dogs: A Systematic Review and Data Synthesis. Prev. Vet. Med. 2022, 209, 105792.
- Pratelli, A.; Buonavoglia, A.; Lanave, G.; Tempesta, M.; Camero, M.; Martella, V.; Decaro, N. One World, One Health, One Virology of the Mysterious Labyrinth of Coronaviruses: The Canine Coronavirus Affair. Lancet Microbe 2021, 2, e646–e647.
- Binn, L.N.; Lazar, E.C.; Keenan, K.P.; Huxsoll, D.L.; Marchwicki, R.H.; Strano, A.J. Recovery and Characterization of a Coronavirus from Military Dogs with Diarrhea. In Proceedings: Annual Meeting; United States Animal Health Association: St. Joseph, MO, USA, 1974; pp. 359–366.
- He, H.-J.; Zhang, W.; Liang, J.; Lu, M.; Wang, R.; Li, G.; He, J.-W.; Chen, J.; Chen, J.; Xing, G.; et al. Etiology and Genetic Evolution of Canine Coronavirus Circulating in Five Provinces of China, during 2018–2019. Microb. Pathog. 2020, 145, 104209.
- Wang, X.; Li, C.; Guo, D.; Wang, X.; Wei, S.; Geng, Y.; Wang, E.; Wang, Z.; Zhao, X.; Su, M.; et al. Co-Circulation of Canine Coronavirus I and IIa/b with High Prevalence and Genetic Diversity in Heilongjiang Province, Northeast China. PLoS ONE 2016, 11, e0146975.
- Pratelli, A.; Tempesta, M.; Elia, G.; Martella, V.; Decaro, N.; Buonavoglia, C. The Knotty Biology of Canine Coronavirus: A Worrying Model of Coronaviruses’ Danger. Res. Vet. Sci. 2022, 144, 190–195.
- Pratelli, A.; Martella, V.; Elia, G.; Decaro, N.; Aliberti, A.; Buonavoglia, D.; Tempesta, M.; Buonavoglia, C. Variation of the Sequence in the Gene Encoding for Transmembrane Protein M of Canine Coronavirus (CCV). Mol. Cell. Probes 2001, 15, 229–233.
- Pratelli, A.; Martella, V.; Pistello, M.; Elia, G.; Decaro, N.; Buonavoglia, D.; Camero, M.; Tempesta, M.; Buonavoglia, C. Identification of Coronaviruses in Dogs That Segregate Separately from the Canine Coronavirus Genotype. J. Virol. Methods 2003, 107, 213–222.
- Pratelli, A.; Martella, V.; Decaro, N.; Tinelli, A.; Camero, M.; Cirone, F.; Elia, G.; Cavalli, A.; Corrente, M.; Greco, G.; et al. Genetic Diversity of a Canine Coronavirus Detected in Pups with Diarrhoea in Italy. J. Virol. Methods 2003, 110, 9–17.
- Lorusso, A.; Decaro, N.; Schellen, P.; Rottier, P.J.M.; Buonavoglia, C.; Haijema, B.-J.; de Groot, R.J. Gain, Preservation, and Loss of a Group 1a Coronavirus Accessory Glycoprotein. J. Virol. 2008, 82, 10312–10317.
- Yesilbag, K.; Yilmaz, Z.; Torun, S.; Pratelli, A. Canine Coronavirus Infection in Turkish Dog Population. J. Vet. Med. Ser. B 2004, 51, 353–355.
- Decaro, N.; Martella, V.; Ricci, D.; Elia, G.; Desario, C.; Campolo, M.; Cavaliere, N.; di Trani, L.; Tempesta, M.; Buonavoglia, C. Genotype-Specific Fluorogenic RT-PCR Assays for the Detection and Quantitation of Canine Coronavirus Type I and Type II RNA in Faecal Samples of Dogs. J. Virol. Methods 2005, 130, 72–78.
- Wesley, R.D. The S Gene of Canine Coronavirus, Strain UCD-1, Is More Closely Related to the S Gene of Transmissible Gastroenteritis Virus than to That of Feline Infectious Peritonitis Virus. Virus Res. 1999, 61, 145–152.
- Naylor, M.J.; Harrison, G.A.; Monckton, R.P.; McOrist, S.; Lehrbach, P.R.; Deane, E.M. Identification of Canine Coronavirus Strains from Feces by S Gene Nested PCR and Molecular Characterization of a New Australian Isolate. J. Clin. Microbiol. 2001, 39, 1036–1041.
- Sanchez-Morgado, J.M.; Poynter, S.; Morris, T.H. Molecular Characterization of a Virulent Canine Coronavirus BGF Strain. Virus Res. 2004, 104, 27–31.
- Escutenaire, S.; Isaksson, M.; Renström, L.H.M.; Klingeborn, B.; Buonavoglia, C.; Berg, M.; Belák, S.; Thorén, P. Characterization of Divergent and Atypical Canine Coronaviruses from Sweden. Arch. Virol. 2007, 152, 1507–1514.
- Decaro, N.; Cordonnier, N.; Demeter, Z.; Egberink, H.; Elia, G.; Grellet, A.; le Poder, S.; Mari, V.; Martella, V.; Ntafis, V.; et al. European Surveillance for Pantropic Canine Coronavirus. J. Clin. Microbiol. 2013, 51, 83–88.
- Buonavoglia, C.; Decaro, N.; Martella, V.; Elia, G.; Campolo, M.; Desario, C.; Castagnaro, M.; Tempesta, M. Canine Coronavirus Highly Pathogenic for Dogs. Emerg. Infect. Dis. 2006, 12, 492–494.
- Pinto, L.D.; Barros, I.N.; Budaszewski, R.F.; Weber, M.N.; Mata, H.; Antunes, J.R.; Boabaid, F.M.; Wouters, A.T.B.; Driemeier, D.; Brandão, P.E.; et al. Characterization of Pantropic Canine Coronavirus from Brazil. Vet. J. 2014, 202, 659–662.
- Alfano, F.; Fusco, G.; Mari, V.; Occhiogrosso, L.; Miletti, G.; Brunetti, R.; Galiero, G.; Desario, C.C.; Cirilli, M.; Decaro, N. Circulation of Pantropic Canine Coronavirus in Autochthonous and Imported Dogs, Italy. Transbound. Emerg. Dis. 2020, 67, 1991–1999.
- Decaro, N.; Martella, V.; Elia, G.; Campolo, M.; Desario, C.; Cirone, F.; Tempesta, M.; Buonavoglia, C. Molecular Characterisation of the Virulent Canine Coronavirus CB/05 Strain. Virus Res. 2007, 125, 54–60.
- Herrewegh, A.A.P.M.; Smeenk, I.; Horzinek, M.C.; Rottier, P.J.M.; de Groot, R.J. Feline Coronavirus Type II Strains 79-1683 and 79-1146 Originate from a Double Recombination between Feline Coronavirus Type I and Canine Coronavirus. J. Virol. 1998, 72, 4508–4514.
- Decaro, N.; Mari, V.; Campolo, M.; Lorusso, A.; Camero, M.; Elia, G.; Martella, V.; Cordioli, P.; Enjuanes, L.; Buonavoglia, C. Recombinant Canine Coronaviruses Related to Transmissible Gastroenteritis Virus of Swine Are Circulating in Dogs. J. Virol. 2009, 83, 1532–1537.
- Decaro, N.; Mari, V.; Elia, G.; Addie, D.D.; Camero, M.; Lucente, M.S.; Martella, V.; Buonavoglia, C. Recombinant Canine Coronaviruses in Dogs, Europe. Emerg. Infect. Dis. 2010, 16, 41–47.
- Regan, A.D.; Millet, J.K.; Tse, L.P.v.; Chillag, Z.; Rinaldi, V.D.; Licitra, B.N.; Dubovi, E.J.; Town, C.D.; Whittaker, G.R. Characterization of a Recombinant Canine Coronavirus with a Distinct Receptor-Binding (S1) Domain. Virology 2012, 430, 90–99.
- Corapi, W.v.; Olsen, C.W.; Scott, F.W. Monoclonal Antibody Analysis of Neutralization and Antibody-Dependent Enhancement of Feline Infectious Peritonitis Virus. J. Virol. 1992, 66, 6695–6705.
- Sha, X.; Li, Y.; Huang, J.; Zhou, Q.; Song, X.; Zhang, B. Detection and Molecular Characteristics of Canine Coronavirus in Chengdu City, Southwest China from 2020 to 2021. Microb. Pathog. 2022, 166, 105548.
More
