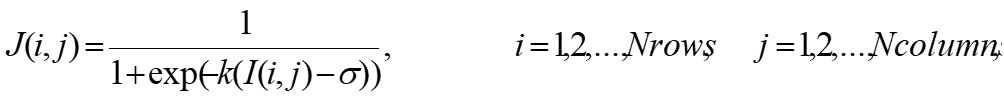Increasing evidence implicates mitochondrial dysfunction in the etiology of Parkinson’s disease (PD). Mitochondrial DNA (mtDNA) mutations represent a possible cause and this mechanism might be shared with the aging process and with other age-related neurodegenerative disorders such as Alzheimer’s disease (AD). We have recently proposed a computerized method for mutated mtDNA characterization able to discriminate between AD and aging. Dealing with mtDNA mutation-based profiling of neurodegenerative disease(s), peripheral blood mtDNA sequences from late-onset PD patients and age-matched controls were analyzed and compared to the revised Cambridge Reference Sequence (rCRS). The chaos game representation (CGR) method, modified to visualize heteroplasmic mutations, was used to display fractal properties of mtDNA sequences and fractal lacunarity analysis was applied to quantitatively characterize PD based on mtDNA mutations. Parameter β, from the hyperbola model function of our lacunarity method, was statistically different between PD and control groups when comparing mtDNA sequence frames corresponding to GenBank np 5713-9713. Our original method, based on CGR and lacunarity analysis, represents a useful tool to analyze mtDNA mutations. Lacunarity parameter β is able to characterize individual mutation profile of mitochondrial genome and could represent a promising index to discriminate between PD and aging.
- Aging
- Biocomplexity
- Chaos Game Representation
- mtDNA
- Fractal Lacunarity
- Parkinson's Disease
1. Introduction
1. Introduction
We have recently proposed a computerized method for mutated mtDNA characterization able to discriminate between AD and aging [1]. Dealing with mtDNA mutations involvement in mitochondrial dysfunction, a mechanism shared by neurodegenerative disorder(s) and aging process, profiling of PD mutated mtDNA has been considered. In this study, peripheral blood mtDNA sequences from late-onset PD patients and age-matched controls were analyzed and compared to the revised Cambridge Reference Sequence (rCRS). CGR method has been used to display DNA fractal properties [2] as modified to visualize heteroplasmic mutations peculiar to mtDNA [1]. Parameter β, from our fractal lacunarity method based on hyperbola model function [3-6], is the measure used to quantitatively characterize PD on the basis of mutated mtDNA. Lacunarity parameter β, in this case, represents a holistic estimate of mtDNA changes comprehensive of number, type, and dislocation of mutation(s), the combination of which could be responsible of disease onset and progression.
We have recently proposed a computerized method for mutated mtDNA characterization able to discriminate between AD and aging [1]. Dealing with mtDNA mutations involvement in mitochondrial dysfunction, a mechanism shared by neurodegenerative disorder(s) and aging process, profiling of PD mutated mtDNA has been considered. In this study, peripheral blood mtDNA sequences from late-onset PD patients and age-matched controls were analyzed and compared to the revised Cambridge Reference Sequence (rCRS). CGR method has been used to display DNA fractal properties [2] as modified to visualize heteroplasmic mutations peculiar to mtDNA [1]. Parameter β, from our fractal lacunarity method based on hyperbola model function [3][4][5][6], is the measure used to quantitatively characterize PD on the basis of mutated mtDNA. Lacunarity parameter β, in this case, represents a holistic estimate of mtDNA changes comprehensive of number, type, and dislocation of mutation(s), the combination of which could be responsible of disease onset and progression.
1.1 Parkinson’s disease and mitochondrial dysfunction
1.1. Parkinson’s disease and mitochondrial dysfunction
Parkinson’s disease (PD) is a neurodegenerative disorder with heavily age-dependent prevalence affecting 1% of over 65s and more than 4% of over 85s subjects worldwide [7]. Pathological characteristics of PD are a loss of dopaminergic neurons and the presence of Lewy bodies (aggregates of α-synuclein and other proteins) in the substantia nigra. Clinical manifestation of the disease is a severe motor dysfunction resulting in resting tremor, rigidity, bradykinesia, and impaired balance. Its causes are still unknown in spite of intense research efforts over many years. Several mechanisms have been proposed to explain cell death in PD, such as oxidative stress, mitochondrial dysfunction, apoptosis, excitotoxicity, and inflammatory responses [8].
Parkinson’s disease (PD) is a neurodegenerative disorder with heavily age-dependent prevalence affecting 1% of over 65s and more than 4% of over 85s subjects worldwide [7]. Pathological characteristics of PD are a loss of dopaminergic neurons and the presence of Lewy bodies (aggregates of α-synuclein and other proteins) in the substantia nigra. Clinical manifestation of the disease is a severe motor dysfunction resulting in resting tremor, rigidity, bradykinesia, and impaired balance. Its causes are still unknown in spite of intense research efforts over many years. Several mechanisms have been proposed to explain cell death in PD, such as oxidative stress, mitochondrial dysfunction, apoptosis, excitotoxicity, and inflammatory responses [8].
Increasing evidence implicates mitochondrial dysfunction in the etiology of PD. This disorder is characterized by systemic loss of mitochondrial complex I activity [9] and disruption of the mechanisms underlying mitophagy [10]. Although the vast majority of PD cases are sporadic, rare familial forms have been reported, with mutations mainly located in α-synuclein, DJ-1, parkin, PTEN-induced kinase 1 (PINK1), and leucinerich repeat kinase 2 (LRRK2) nuclear genes [11]. These mutated genes are all involved in mitochondrial physiology suggesting that altered mitochondrial function could cause PD. Much evidence suggests that altered mitochondrial function in sporadic PD cases could be related to alterations of mitochondrial DNA (mtDNA).
Increasing evidence implicates mitochondrial dysfunction in the etiology of PD. This disorder is characterized by systemic loss of mitochondrial complex I activity [9] and disruption of the mechanisms underlying mitophagy [10]. Although the vast majority of PD cases are sporadic, rare familial forms have been reported, with mutations mainly located in α-synuclein, DJ-1, parkin, PTEN-induced kinase 1 (PINK1), and leucinerich repeat kinase 2 (LRRK2) nuclear genes [11]. These mutated genes are all involved in mitochondrial physiology suggesting that altered mitochondrial function could cause PD. Much evidence suggests that altered mitochondrial function in sporadic PD cases could be related to alterations of mitochondrial DNA (mtDNA).
Three classes of mtDNA alterations are implicated: ancient maternally inherited polymorphisms, recent maternally inherited pathogenic mutations, and somatic mtDNA mutations [12,13]. These last mutations accumulate in post-mitotic cells with age and amplify the biochemical effects of the former two classes. Accumulation of somatic mtDNA mutations has been shown to be an important factor in the development of PD and Alzheimer’s disease (AD) as well as in aging [14]. The role of mtDNA in PD is supported by the analysis of cybrid cells, generated by the fusion of platelets (from control or patient) as mitochondria donor with recipient cells deprived of mitochondria. The common nuclear background of cybrids allows excluding the nuclear genome effect in the bioenergetics characterization. PD cybrids show depolarized mitochondria, reduced Complex I (CI) activity, increased ROS (reactive oxygen species) production and lower ATP production, most hallmarks of PD [15].
Three classes of mtDNA alterations are implicated: ancient maternally inherited polymorphisms, recent maternally inherited pathogenic mutations, and somatic mtDNA mutations [12][13]. These last mutations accumulate in post-mitotic cells with age and amplify the biochemical effects of the former two classes. Accumulation of somatic mtDNA mutations has been shown to be an important factor in the development of PD and Alzheimer’s disease (AD) as well as in aging [14]. The role of mtDNA in PD is supported by the analysis of cybrid cells, generated by the fusion of platelets (from control or patient) as mitochondria donor with recipient cells deprived of mitochondria. The common nuclear background of cybrids allows excluding the nuclear genome effect in the bioenergetics characterization. PD cybrids show depolarized mitochondria, reduced Complex I (CI) activity, increased ROS (reactive oxygen species) production and lower ATP production, most hallmarks of PD [15].
1.2 Biocomplexity, chaos and fractality in aging and disease
1.2. Biocomplexity, chaos and fractality in aging and disease
Paradigms, such as theory of complexity, chaos, and fractals, suggest new approaches and provide new tools to the study of aging processes. In particular, the concept that aging can be considered as a “secondary product” of the temporal evolution of a dynamic nonlinear system [16-18], governed by the laws of deterministic chaos, can explain the heterogeneity of the senescent phenotype [1,18]. The concept that a complex system with a chaotic behavior often generates fractals [19] highlights the usefulness of fractal analysis as a suitable tool to measure biocomplexity and its changes with aging at both functional and structural levels [20-22].
Paradigms, such as theory of complexity, chaos, and fractals, suggest new approaches and provide new tools to the study of aging processes. In particular, the concept that aging can be considered as a “secondary product” of the temporal evolution of a dynamic nonlinear system [16][17][18], governed by the laws of deterministic chaos, can explain the heterogeneity of the senescent phenotype [1][18]. The concept that a complex system with a chaotic behavior often generates fractals [19] highlights the usefulness of fractal analysis as a suitable tool to measure biocomplexity and its changes with aging at both functional and structural levels [20][21][22].
Fractal analysis can measure variations of complexity in biosystems that evolve with time by following different trajectories. Individual specific genetic-environment interactions define the senescent phenotype as normal aging, pathological aging, or successful aging [1,18]. Fractal analysis, therefore, represents a promising tool to give insight into the search of good biomarkers useful to discriminate between physiological and pathological aging as well as between age-related and age-associated diseases.
Fractal analysis can measure variations of complexity in biosystems that evolve with time by following different trajectories. Individual specific genetic-environment interactions define the senescent phenotype as normal aging, pathological aging, or successful aging [1][18]. Fractal analysis, therefore, represents a promising tool to give insight into the search of good biomarkers useful to discriminate between physiological and pathological aging as well as between age-related and age-associated diseases.
Fractal dimension (FD) has been widely used to measure complexity variation of most biomedical functions and structures in aging and disease. Fractal lacunarity, another fractal property, describes the texture and measures fractal space filling capacity of a fractal [23]. The term lacunarity (from Latin lacuna, lack or hole) was coined by Mandelbrot by referring to the gap distribution in a fractal, and lacunarity analysis was initially introduced to differentiate fractal objects displaying the same FD but having a very different appearance [23].
Fractal dimension (FD) has been widely used to measure complexity variation of most biomedical functions and structures in aging and disease. Fractal lacunarity, another fractal property, describes the texture and measures fractal space filling capacity of a fractal [23]. The term lacunarity (from Latin lacuna, lack or hole) was coined by Mandelbrot by referring to the gap distribution in a fractal, and lacunarity analysis was initially introduced to differentiate fractal objects displaying the same FD but having a very different appearance [23].
We have recently proposed a method of lacunarity analysis in chaos game representation (CGR) images of mtDNA sequences [1] able to discriminate between aging and AD on the basis of mtDNA mutation profiles. The method was developed taking into account the complexity of living beings and fractal properties of many anatomic and physiologic structures, among which is mtDNA [24-26]. Fractal lacunarity analysis was chosen to develop our method. In fact, lacunarity analysis has been also introduced as a more general technique able to describe both random and fractal spatial patterns [27-28], thus, overcoming the limits of fractal analysis applied to natural objects [18,23,28]. In addition, dealing with mtDNA mutations as gaps in the nucleotide sequence, fractal lacunarity appears a suitable tool to differentiate between aging and neurodegenerative disease(s).
We have recently proposed a method of lacunarity analysis in chaos game representation (CGR) images of mtDNA sequences [1] able to discriminate between aging and AD on the basis of mtDNA mutation profiles. The method was developed taking into account the complexity of living beings and fractal properties of many anatomic and physiologic structures, among which is mtDNA [24][25][26]. Fractal lacunarity analysis was chosen to develop our method. In fact, lacunarity analysis has been also introduced as a more general technique able to describe both random and fractal spatial patterns [27][28], thus, overcoming the limits of fractal analysis applied to natural objects [18][23][28]. In addition, dealing with mtDNA mutations as gaps in the nucleotide sequence, fractal lacunarity appears a suitable tool to differentiate between aging and neurodegenerative disease(s).
2. Outline of the method
2. Outline of the method
for mtDNA profiling
2.1 Chaos Game representation of mtDNA
2.1. Chaos Game representation of mtDNA
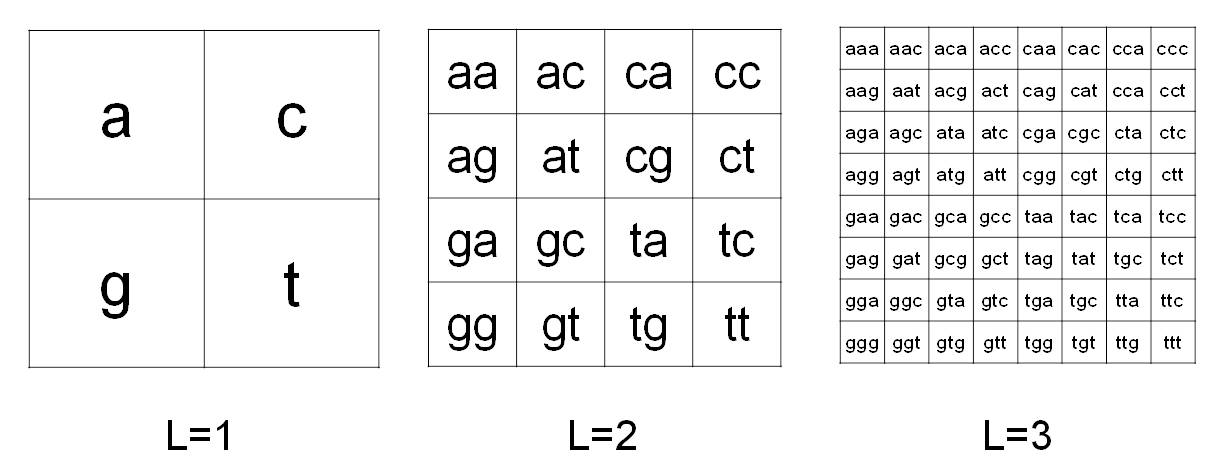
CGR was proposed by Jeffrey [2] to codify in images the information contained in the DNA. The CGR method as modified in [1] is used to analyze the sequence of mtDNA from Affymetrix MitoChips. In particular, let
CGR was proposed by Jeffrey [2] to codify in images the information contained in the DNA. The CGR method as modified in [1] is used to analyze the sequence of mtDNA from Affymetrix MitoChips. In particular, let
S
be a finite string given by a DNA sequence, where the alphabet is {a, c, g, t}. The CGR image of
S
is obtained by the frequency occurrence of all the possible substrings of
S
having fixed length
L
. Due to the four-symbol alphabet, the substrings of S can be organized in a square matrix of order 2
L
. Figure 1 shows a pictorial description of this organization for
L=1,2,3
. From this relation, a position
p(s)
is associated to each possible substring
s
of
S
with length
L
. Then, the CGR matrix of
S
is computed by the following rule: the value of the entry at position
p(s)
is given by the frequency of
s
in
S
.
Figure 1
. Chaos Game Representation method. CGR organization in matrices 2
L
x2
L
for L=1,2,3 in the case of four- symbol alphabet {a,c,g,t}.
The proposed modification [1] allows us to deal with undetermined mtDNA typing symbols (heteroplasmic mutations). More precisely, substrings
The proposed modification [1] allows us to deal with undetermined mtDNA typing symbols (heteroplasmic mutations). More precisely, substrings
s
containing undetermined symbols are formally substituted by the corresponding multiple strings obtained by solving such undetermined symbols and each one of the resulting strings has a fractional weight
w
in the frequency computation.
The following examples show the generation procedure for the substrings containing undetermined symbols and the determination of the weight in the frequency computation.
Example 1
.
Let L=5 and s=’tamcg’, where the undetermined symbol ‘m’ means ‘a’ or ‘c’. This string is substituted by s1=’taacg’ and s2=’taccg’; the weight is w=1/2.
Example 2
.
Let L=6 and s=’tavcgm’, where ‘v’ means ‘a’, ‘c’, or ‘g’. This string is substituted by s1=’taacga’, s2=’taccga’, s3=’tagcga’, s4=’taacgc’, s5=’taccgc’, s6=’tagcgc’; the weight is w=1/6.
In the modified version, the CGR matrix is defined in the same way, where the frequency of each substring
s
of
S
is computed by using the weights
w
and assuming
w
=1 for substrings having no undetermined symbols.
This algorithm has been implemented in a MATLAB program (the MatWorks, Inc.) that, also, provides a report with additional information on mtDNA sequence processed, i.e. type and number of nucleotide(s), number and position of homoplasmic/heteroplasmic mutations.
2.2. Fractal Lacunarity of mtDNA in CGR
2.2 Fractal Lacunarity of mtDNA in CGR
Fractal lacunarity analysis of CGR matrix has been performed by using the gliding box algorithm (GBA), see [27] for a detailed description. This method has been applied to several medical investigations and it is already described in [3][4] and in [5][6] for a modified version. In this section, GBA is sketched out for the convenience of the reader.
Fractal lacunarity analysis of CGR matrix has been performed by using the gliding box algorithm (GBA), see [27] for a detailed description. This method has been applied to several medical investigations and it is already described in [3,4] and in [5,6] for a modified version. In this section, GBA is sketched out for the convenience of the reader.
GBA is based on the analysis of the mass distribution in a given set, where the set is associated to the CGR matrix and the mass is associated to the total frequency obtained in the entries of CGR matrix. In particular, GBA computes the lacunarity function Λ(b) as the ratio between the moments of order 1 and 2 of the box mass, within a box of size b>0 moving on the set one space unit at a time. This algorithm can be adapted to deal with gray scale images and provides a simple extension of the method for binary images, see Zaia et al. [5][6] for details. The efficiency of such an algorithm is usually improved by a pre-processing step, where the original image I is used to compute a refined version J as follows:
GBA is based on the analysis of the mass distribution in a given set, where the set is associated to the CGR matrix and the mass is associated to the total frequency obtained in the entries of CGR matrix. In particular, GBA computes the lacunarity function Λ(b) as the ratio between the moments of order 1 and 2 of the box mass, within a box of size b>0 moving on the set one space unit at a time. This algorithm can be adapted to deal with gray scale images and provides a simple extension of the method for binary images, see Zaia et al. [5,6] for details. The efficiency of such an algorithm is usually improved by a pre-processing step, where the original image I is used to compute a refined version J as follows:
|
|
(1) |
and k, σ > 0 are two given parameters. It is worth noting that the procedure goes toward a complete binarization by increasing parameter k, related to sigmoid regularization.
The GBA method has been implemented in a Matlab program that calculates the values of lacunarity Λ(b), for each integer value of b between b
min
and b
max
, where b
min
, b
max
are given integer multiples of the pixel size in the image under consideration. Once the lacunarity function Λ(b), b=b
min
, b
min
+1,…, b
max
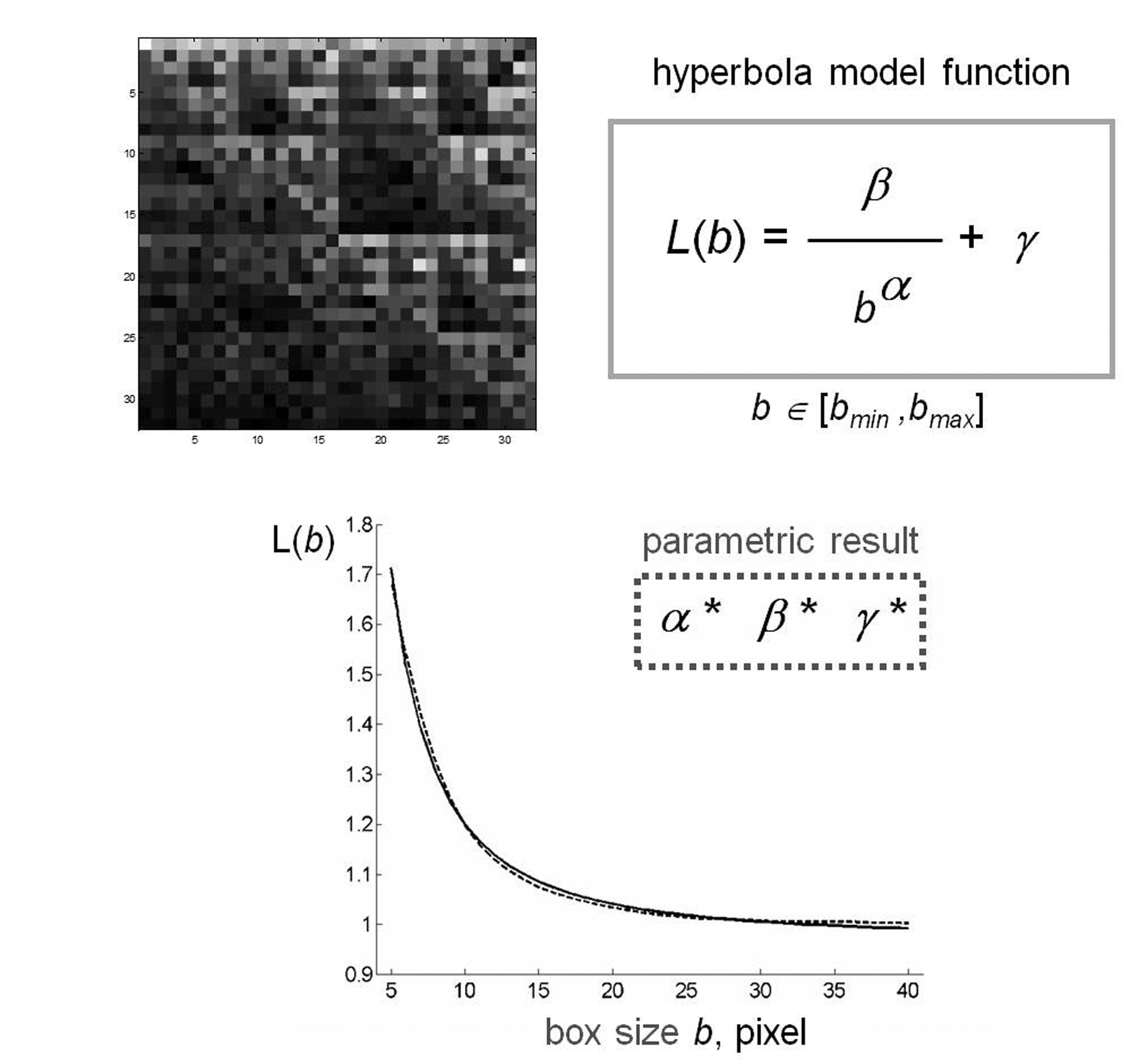
is obtained, the program shows the results on a graph and computes the least squared best-fit through the data by using the following model:
|
|
PD patients |
Controls |
|
|
Number (M/F) |
15 (7/8) |
15 (6/9) |
|
Age (year) |
78.8 ± 6.0 |
80.8 ± 5.1 |
|
H&Y |
2.5 (2.0-3.0) |
|
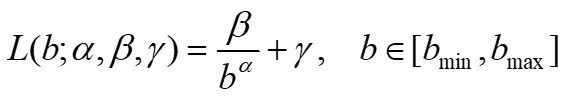 |
(2) |
where α, β, γ are suitable parameters. We note that parameter α is related to the fractal dimension of the set and parameter β characterizes the lacunarity of the set [4][5].
where α, β, γ are suitable parameters. We note that parameter α is related to the fractal dimension of the set and parameter β characterizes the lacunarity of the set [4,5].
Figure 2
. Schematic representation of fractal lacunarity analysis. (Top left) rCRS mtDNA image generated by CGR matrix for L=5 is a quare. The plot (bottom) represents the result of GBA application (dotted line), for bmin=3, as fitted by hyperbola function (solid line) used to calculate the triplet of parameters α, β, γ.
Figure 2 shows a schematic representation of the method applied to a CGR matrix generated by rCRS. CGR matrices generated from different mtDNA sequences produced a similar curvilinear plot. The almost perfect overlap of the two experimental and theoretic curves supports the appropriateness of our choice of hyperbola model function to fit the gliding box curve.
3. Results
Thirty mtDNA sequences from peripheral blood of 15 late-onset PD patients and 15 age- and sex-matched controls (Table 1) were processed to generate CGR matrices for L=5 and L=6. Any set of matrices 2
3. Results
Thirty mtDNA sequences from peripheral blood of 15 late-onset PD patients and 15 age- and sex-matched controls (Table 1) were processed to generate CGR matrices for L=5 and L=6. Any set of matrices 2
L
x2
L
was analyzed to verify the potential of our lacunarity parameter β in characterizing alterations of mtDNA in aging and PD, based on the revised Cambridge Reference Sequence (rCRS).
Table 1. Characteristics of subjects included in the study.
Characteristics of subjects included in the study.
|
CPS |
2.0 (0.5-3.0) |
2.0 (0.5-2.0) |
|
ADL |
1.0 (0.0-2.0) |
0.0 (0.0-3.0) |
Values are expressed as mean ± SD or median (interquartile range) in the case of non-normally distributed variables; PD: Parkinson’s Disease; H&Y: Hoehn & Yahr stage; CPS: Cognitive Performance Scale; ADL: Activities of Daily Living hierarchy scale.
3.1 Chaos Game Representation
-
- 2004
- ,
- 41
- , 41–62, doi:10.1385/CBB:41:1:041.
- Hao, B.L. Fractals from genomes—Exact solutions of a biology-inspired problem.
- Physica A
-
- 2000
- ,
- 282
- , 225–246, doi:10.1103/PhysRevLett.84.386.
- Cattani, C.; Pierro, G. On the fractal geometry of DNA by the binary image analysis.
- Math. Biol.
-
- 2013
- ,
- 75
- , 1544–1570, doi:10.1007/s11538-013-9859-9.
- Allain, C.; Cloitre, M. Characterizing the lacunarity of random and deterministic fractal sets.
- Rev. A
-
- 1991
- ,
- 44
- , 3552–3558, doi:10.1103/physreva.44.3552.
- Plotnick, R.E.; Gardner, R.H.; Hargrove, W.W.; Prestegard, K.; Perlmutter, M. Lacunarity analysis: A general technique for the analysis of spatial patterns.
- Rev. E
-
- 1996
- ,
- 53
- , 5461–5468, doi:10.1103/physreve.53.5461.
- Wang, Y.; Hill, K.; Singh, S.; Kari, L. The spectrum of genomic signatures: From dinucleotides to chaos game representation. Gene 2005, 346, 173–185, doi:10.1016/j.gene.2004.10.021.
- Casoli,T.; Lisa, R.; Fabbietti, P.; Conti, F. Analysis of mitochondrial DNA allelic changes in Parkinson’s disease: A preliminary study. Aging Clin. Exp. Res. 2020, 32, 345–349, doi:10.1007/s40520-019-01197-4.
- Casoli, T.; Spazzafumo, L.; Di Stefano, G.; Conti, F. Role of diffuse low-level heteroplasmy of mitochondrial DNA in Alzheimer’s disease neurodegeneration. Front. Aging Neurosci. 2015, 7, 142, doi:10.3389/fnagi.2015.00142.
- Coskun, P.; Wyrembak, J.; Schriner, S.E.; Chen, H.W.; Marciniack, C.; Laferla, F.; Wallace, D.C. A mitochondrial etiology of Alzheimer and Parkinson disease. Biophys. Acta 2012, 1820, 553–564, doi:10.1016/j.bbagen.2011.08.008.
- Dhillon, V.S.; Fenech, M. Mutations that affect mitochondrial functions and their association with neurodegenerative diseases. Res. Rev. Mutat. Res. 2014, 759, 1–13, doi:10.1016/j.mrrev.2013.09.001.
- Smigrodzki, R.; Parks, J.; Parker, W.D. High frequency of mitochondrial complex I mutations in Parkinson’s disease and aging. Aging 2004, 25, 1273–1281, doi:10.1016/j.neurobiolaging.2004.02.020.
- Ikebe, S.; Tanaka, M.; Ohno, K.; Sato, W.; Hattori, K.; Kondo, T.; Mizuno, Y.; Ozawa, T. Increase of deleted mitochondrial DNA in the striatum in Parkinson’s disease and senescence. Biophys. Res. Commun. 1990, 170, 1044–1048, doi:10.1016/0006-291X(90)90497-B.
- Schapira, A.H.; Holt, I.J.; Sweeney, M.; Harding, A.E.; Jenner, P.; Marsden, C.D. Mitochondrial DNA analysis in Parkinson’s disease. Disord. 1990, 5, 294–247, doi:10.1002/mds.870050406.
- Lestienne, P.; Nelson, I.; Riederer, P.; Reichmann, H.; Jellinger, K. Mitochondrial DNA in postmortem brain from patients with Parkinson’s disease. Neurochem. 1991, 56, 1819–1819, doi:10.1111/j.1471-4159.1991.tb02087.x.
- Kraytsberg, Y.; Kudryavtseva, E.; McKee, A.C.; Geula, C.; Kowall, N.W.; Khrapko, K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Genet. 2006, 38, 518–520, doi:10.1038/ng1778.
- Bender, A.; Krishnan, K.J.; Morris, C.M.; Taylor, G.A.; Reeve, A.K.; Perry, R.H.; Jaros, E.; Hersheson, J.S.; Betts, J.; Klopstock, T.; et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Genet. 2006, 38, 515–517, doi:10.1038/ng1769.
- Hua, F.; Zhang, X.; Hou, B.; Xue, L.; Xie, A. Relationship between mitochondrial DNA A10398G polymorphism and Parkinson’s disease: A meta-analysis. Oncotarget 2017
|
|
PD patients |
Controls |
|
Number (M/F) |
15 (7/8) |
15 (6/9) |
|
Age (year) |
78.8 ± 6.0 |
80.8 ± 5.1 |
|
H&Y |
2.5 (2.0-3.0) |
|
|
CPS |
2.0 (0.5-3.0) |
2.0 (0.5-2.0) |
|
ADL |
1.0 (0.0-2.0) |
0.0 (0.0-3.0) |
Values are expressed as mean ± SD or median (interquartile range) in the case of non-normally distributed variables; PD: Parkinson’s Disease; H&Y: Hoehn & Yahr stage; CPS: Cognitive Performance Scale; ADL: Activities of Daily Living hierarchy scale.
3.1. Chaos Game Representation
Figure 3 shows a set of six CGR images generated from rCRS with matrices 2
Figure 3 shows a set of six CGR images generated from rCRS with matrices 2
L
x2
L
for L=1 to L=6. Note that fractal structure of human mtDNA resembles the Sierpinski triangle (Figure 3b). The same Sierpinski-like structure for human mtDNA was previously reported by Wang et al. [29]. It differs from human nuclear DNA and from DNA of other species, thus highlighting a species- and type-specificity of DNA fractal representation.
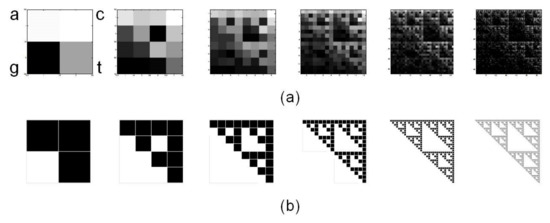
Figure 3.
Chaos Game Representation (CGR) of human mtDNA. (a) CGR matrices for L=1 to L=6 of whole revised Cambridge Reference Sequence, rCRS, display self-similarity, a property peculiar to fractals, resembling (b) the triangle of Sierpinski, an ideal fractal built through repeated iterations starting from a square.
Table 2 summarizes results on mtDNA sequences from the whole sample of 30 subjects. In particular, both PD and control mtDNA sequences show decreased numbers of the four nucleotide bases when compared to rCRS. These changes, accounting for similar numbers of no-call, homoplasmic, and heteroplasmic mutations observed in PD and control subjects, do not show statistically significant differences.
Table 2.
Characteristics of mtDNA sequences processed by the proposed method.
- ,
- 8
- , 78023–78030, doi:10.18632/oncotarget.209207.
- Coxhead, J.; Kurzawa-Akanbi, M.; Hussain, R.; Pyle, A.; Chinnery, P.; Hudson, G. Somatic mtDNA variation is an important component of Parkinson’s disease.
- Aging
-
- 2016
- ,
- 38
- , 217e1–217e6, doi:10.1016/j.neurobiolaging.2015.10.036.
- Lin, M.T.; Cantuti-Castelvetri, I.; Zheng, K.; Jackson, K.E.; Tan, Y.B.; Arzberger, T.; Lees, A.J.; Betensky, R.A.; Beal, M.F.; Simon, D.K. Somatic mitochondrial DNA mutations in early Parkinson and incidental Lewy body disease.
- Neurol.
-
- 2012
- ,
- 71
- , 850–854, doi:10.1002/ana.23568.
- Wallace, D.C. Why do we have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu. Rev. Biochem.
- 2007
- , 76, 781–821. Annu. Rev. Biochem. 2007, 76, 781–821, doi:10.1146/annurev.biochem.76.081205.150955.
- Lipsitz, L.A. Physiological complexity, aging, and the path to frailty.
- Aging Knowl. Environ.
-
- 2004
- ,
- 16
- , pe16, doi:10.1126/sageke.2004.16.pe16.
- Postuma, R.B.; Berg, D.; Stern, M.; Poewe, W.; Olanow, C.W.; Oertel, W.; Obeso, J.; Marek, K.; Litvan, I.; Lang, A.E.; et al. MDS clinical diagnostic criteria for Parkinson’s disease.
- Disord.
-
- 2015
- ,
- 30
- , 1591–1601, doi:10.1002/mds.26424.
- Broderick, P.A.; Wenning, L. Neuromolecular imaging in Parkinson’s disease. In
- Compendium on Parkinson’s Disease; Preedy, V.R
- .; Elsevier Press: London, UK, 2020; Chapter 16.
|
Number |
rCRS |
PD patients |
Controls |
p Value |
|
Subjects |
|
15 |
15 |
|
|
Adenine |
5117 |
4884 ± 252 |
4912 ± 110 |
0.359 |
|
Cytosine |
5175 |
4658 ± 206 |
4618 ± 168 |
0.301 |
|
Guanine |
2163 |
2102 ± 92 |
2108 ± 49 |
0.421 |
|
Thymine |
4089 |
3880 ± 239 |
3916 ± 101 |
0.311 |
|
No-call |
- |
1009 ± 765 |
980 ± 390 |
0.453 |
|
Homoplasmy |
- |
18 ± 8 |
21 ± 8 |
0.196 |
|
Heteroplasmy |
- |
12 ± 13 |
10 ± 6 |
0.332 |
Values are expressed as mean ± SD; p Values have been calculated by one-tailed t-test for p≤0.05 to compare differences between PD and control groups; rCRS: revised Cambridge Reference Sequence; PD: Parkinson’s Disease.
3.2. Fractal Analysis of mtDNA in Aging and Parkinson’s Disease
3.2 Fractal Analysis of mtDNA in Aging and Parkinson’s Disease
Our method of fractal lacunarity analysis was systematically applied to CGR matrices 2
L
x2
L
generated from the above described mtDNA sequences for L=5 (32x32) and L=6 (64x64) based on results obtained in a previous study on mtDNA in AD [1]
.
For the best characterization of any mtDNA sequence analyzed, the combination of coefficients, both sigmoid coefficients (k and σ) and b
min
, was confirmed to be: k equal to 7 and σ equal to 0.7; b
min
equal to 3 and 5. For this set of coefficients, we obtained comparable results related to rCRS.
Examples of CGR images for L=5 from whole mtDNA sequence of rCRS, a PD patient, and an age-matched control are reported in Figure 4a. In spite of a similar display among the three kinds of mtDNA sequences, parameter b values related to both PD and control subjects significantly differ from rCRS. In particular, lower b values, observed in PD and control mtDNA when compared to rCRS, correspond to the degree of alterations (number, type and dislocation) of the nucleotide sequences considered. In this study, however, we didn’t find any statistically significant difference between PD and control groups for both b
min
3 and 5 in matrices 32x32 (L=5) and 64x64 (L=6). Other matrix sizes (i.e., L=4, L=7, L=8) tested did not show statistical differences as well. We also noted that two PD subjects and three controls did not exhibit the characteristic hyperbola-like lacunarity curve of their mtDNA sequence CGR for L=5 and b
min
=3.
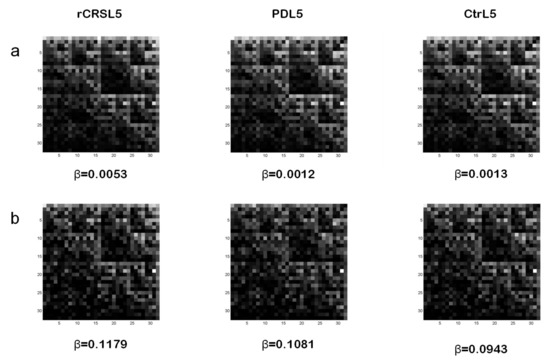
Figure 4.
Examples of CGR images of whole mtDNA sequences (
a
) and related GenBank np 5713-9713 frames (
b
) from different subjects. Lacunarity parameter β value for CGR matrices 32x32 (L=5) generated from mtDNA of rCRS (left), a PD patient (middle), and a control (right) is reported. CGR: Chaos Game Representation; rCRS: revised Cambridge Reference Sequence; PD: Parkinson’s Disease.
Because of the low number (58) of np differing between PD and control groups [30] we decided to analyze a shorter frame of mtDNA sequence. In particular, we considered the frame length corresponding to 5713–9713 np in the GenBank. It represents about ¼ of the whole mtDNA sequence and contains 41% (24 out of 58) np with increased presence of non-reference alleles in PD patients. Interestingly, mutations in this frame involve genes, such as COI, COII, COIII of complex IV, and ATPase6 of complex V, of the respiratory chain. These complexes play a very important role in energy production; therefore, their mutations may represent a serious impairment of cell metabolism. Figure 4b shows examples of CGR for L=5 of the mtDNA frame considered, and related β values, from rCRS and from the same PD and control subjects of Figure 4a.
Because of the low number (58) of np differing between PD and control groups [30] we decided to analyze a shorter frame of mtDNA sequence. In particular, we considered the frame length corresponding to 5713–9713 np in the GenBank. It represents about ¼ of the whole mtDNA sequence and contains 41% (24 out of 58) np with increased presence of non-reference alleles in PD patients. Interestingly, mutations in this frame involve genes, such as COI, COII, COIII of complex IV, and ATPase6 of complex V, of the respiratory chain. These complexes play a very important role in energy production; therefore, their mutations may represent a serious impairment of cell metabolism. Figure 4b shows examples of CGR for L=5 of the mtDNA frame considered, and related β values, from rCRS and from the same PD and control subjects of Figure 4a.
Lacunarity analysis of 32x32 CGR matrices generated from this mtDNA frame length showed a significant lower β value in both PD and control groups when compared to rCRS. After exclusion of two PD patients and two controls displaying abnormally high α values, a statistically significant difference was observed between PD patients and age-matched control subjects with lower β values found in controls. Table 3 summarizes lacunarity results from both mtDNA whole and frame lengths for CGR size 32x32 (L=5) and b
min
=3. Randomized sampling method was applied to the sample of subjects under study to confirm that the statistically significant difference between PD and controls is not reached by chance alone. Results from 20-fold repeated randomized sampling into two mixed groups do not show any statistical difference.
Table 3.
Fractal parameters from lacunarity analysis method based on hyperbola model function of mtDNA in Parkinson’s disease.
|
mtDNA
|
whole α β |
frame1 α β |
||
|
rCRS |
1.6375 |
0.0053 |
1.3224 |
0.1179 |
|
|
|
|
|
|
|
PD patients |
1.6405 ± 0.0005 |
0.0011 ± 0.0004 |
1.3106 ± 0.060 |
0.1019 ± 0.061 |
|
Controls |
1.6403 ± 0.0007 |
0.0009 ± 0.0013 |
1.3150 ± 0.073 |
0.0910 ± 0.061 |
|
p Value |
0.179 |
0.151 |
0.433 |
0.028 |
Values are mean ± standard deviation
1
mtDNA sequence frame corresponding to GenBank np 5713-9713.
4. Biocomplexity, Fractality and mtDNA profiling of Parkinson's disease
In this study we confirm that fractal analysis represents a useful tool to discriminate between aging and age-related pathologies [1][6]. In particular, fractal lacunarity analysis of mtDNA, performed by our method, is able to highlight a statistically significant difference of mtDNA mutation profile between PD patients and age-matched controls. Differently from a previous study on AD [1], we found that CGR images of the whole mtDNA sequence from PD patients display fractal properties similar to age-matched controls while parameter β (representative of lacunarity) is significantly lower in both groups when compared to rCRS. Keeping in mind that low β values correspond to high lacunarity, that is high degree of alterations of mtDNA, this result is consistent with previous observations of only 58 statistically different np in PD [30] vs. 270 found in AD [31]. The low number of variants, together with their type and dislocation along the whole mtDNA sequence, is not sufficient to characterize a PD mutation-based profile vs. aging. However, by focusing on a shorter frame of mtDNA sequence, corresponding to GenBank np 5713-9713, that includes 41% of total np whose nucleotide distribution is different between PD and controls, we found a statistically significant difference of lacunarity between PD and control groups with lower β values observed in the last one.
From literature, it is quite accepted that mitochondrial dysfunction and oxidative damage play an important role in the pathogenesis of PD as it is for other neurodegenerative diseases [32][33]. However, the mechanisms involved have not been clearly identified yet. Most mechanisms of mitochondrial dysfunction observed in PD are shared with the aging process [34][35] and overlapping of mtDNA alterations in PD and controls, without a clear-cut characterization of PD mtDNA mutation profile, observed also in this study when dealing with the whole mtDNA sequence, can explain the undefined genotype picture of this disease. As a matter of fact, although more deletions were found in mtDNA of PD patients, comparison with age-matched controls did not show any remarkable increase [35][36]. With the advent of more sensitive techniques to investigate the deleted mtDNA in individual cells, the increased number of mtDNA deletions in individual neurons of the substantia nigra has been confirmed in PD patients older than 65 years [36]. Another study [37], investigating the number of mtDNA deletions in the brains of subjects with parkinsonism, widespread Lewy body deposition, and less severe loss of dopaminergic neurons in substantia nigra, has reported a high proportion of mtDNA deletions (43%) in controls, which increased with age. However, a higher number of mtDNA deletions was observed in patients with parkinsonism and dementia (52%).
Generally, sequencing of mtDNA from PD patients has been performed in unselected groups, with and without a mitochondrial deficiency [38][39]. Although the results from some studies have suggested increased frequency of specific mtDNA polymorphisms in PD patients, this aspect has not been confirmed in all studies. In addition, PD has not been strongly associated with any specific mtDNA mutation yet. The increased frequency of specific mutations or mtDNA haplogroups observed in controls has suggested that they may play a protective role against PD, while some others have been associated with PD. Increased levels of somatic mtDNA point mutations in the substantia nigra have been found also in early PD, suggesting that such mutations occur in the early phase of this pathology [40][41][42]. If mtDNA mutations have any effect on this disease, their role appears to be complex and could involve specific haplotypes or combinations of sequence changes that modify mitochondrial function and make the system more vulnerable to nuclear genetic effects and/or environmental influences [43].
Such a picture highlights the usefulness of the theory of complexity and the laws of chaos to explain and characterize individual phenotypes evolving over time as ‘normal’ aging or pathological aging [1][6]. The inter-individual variability observed in senescent phenotype can be explained in the light of the theory of complexity by considering longevity as a “secondary product” of evolution of a dynamic nonlinear system. In fact, human beings as complex systems are made up of numerous sub-systems (nervous, endocrine, cardiovascular, … systems), interacting with each other. Each sub-system is further subdivided into interacting lower components (organs, tissues, cells, …) and so on at lower levels of organization. The organization into hierarchy and the laws of chaos can explain their evolution, from development to senescence, through the maintenance of a homeostatic dynamic equilibrium of their integrated functions as an adaptive response to continuous noxae from both endogenous and exogenous environments.
Based on this holistic point of view, aging has been defined as the temporal evolution of a complex system that evolves, under the influence of both endogenous and exogenous environments, with loss of complexity during aging [1][12][44]. Human beings, as complex systems characterized by a chaotic behavior, generate fractals that can be observed at both structural and functional levels. Fractal analysis, therefore, represents an intriguing tool useful to measure biocomplexity changes with aging and pathology. Fractal analysis can measure variations of complexity in biosystems that evolve with time by following different trajectories. The specific individual genetic-environment interactions determine the senescent phenotype that evolves as ‘normal’ aging, pathological aging, or successful aging [1][17]. Dealing with mutations of mtDNA as gaps in the nucleotide sequence, fractal lacunarity represents the most suitable tool to differentiate between PD and aging. In fact, lacunarity gives a holistic estimate of changes that occur in mtDNA sequences, comprising of number, type, and dislocation of mutation(s), the combination of which could contribute to PD onset and progression.
It is worth noting that a full certain diagnosis of PD is impossible during life: 75%–95% of PD patients have their diagnosis confirmed only post-mortem [45]. Diagnostic accuracy varies considerably depending on disease duration (lower on first visit), age, clinician expertise, and increasing understanding of this pathology. In this context, advances in live imaging, such as neuromolecular imaging, could improve the understanding as well as diagnosis of PD [46]. Often, failure in recognizing other pathologies causing neurodegenerative disorder or secondary parkinsonism as well as the absence of a true progressive parkinsonian disorder are common causes of diagnostic error [45].
Dealing with this and other observations discussed above, it would be interesting to verify the potential of our original method for mtDNA mutation-based profiling of neurodegenerative disease(s) also for the whole mtDNA sequence from PD patients as per AD [1]. In fact, we do not know whether the positive results observed on the smaller mtDNA frame but not on the whole sequence is peculiar to the disease or, rather, it is attributable to the sample of subjects used in this study. As a matter of fact, PD patients were diagnosed for late-onset PD; therefore, accumulation of mtDNA somatic mutations that occur with aging could mask PD specific point mutations or sequence changes of mtDNA. In addition, taking into account that PD is a neurodegenerative disease characterized by motor dysfunction, it would be useful to compare PD with age-matched controls without altered motor function. Our sample didn’t show any statistically significant difference as far as cognitive and motor function, unrelated to PD dysfunction, are concerned, the only difference accounting for limited head rotation in PD group. Last but not least characteristic of the sample under study deals with hospitalization: both PD patients and age-matched controls were hospitalized for other and different pathologies, probably with or without implication for mitochondrial dysfunction. In this context, it would be interesting to investigate whether the similar mtDNA mutation profile, observed in the whole mtDNA sequence of both PD and controls, could represent a mitochondrial genome profile peculiar to pathological senescent phenotype, characterized by multimorbidity affecting more than 60% elderly people that experience a pathological aging.
4. Biocomplexity, Fractality and mtDNA profiling of Parkinson's disease
In this study we confirm that fractal analysis represents a useful tool to discriminate between aging and age-related pathologies [1,6]. In particular, fractal lacunarity analysis of mtDNA, performed by our method, is able to highlight a statistically significant difference of mtDNA mutation profile between PD patients and age-matched controls. Differently from a previous study on AD [1], we found that CGR images of the whole mtDNA sequence from PD patients display fractal properties similar to age-matched controls while parameter β (representative of lacunarity) is significantly lower in both groups when compared to rCRS. Keeping in mind that low β values correspond to high lacunarity, that is high degree of alterations of mtDNA, this result is consistent with previous observations of only 58 statistically different np in PD [30] vs. 270 found in AD [31]. The low number of variants, together with their type and dislocation along the whole mtDNA sequence, is not sufficient to characterize a PD mutation-based profile vs. aging. However, by focusing on a shorter frame of mtDNA sequence, corresponding to GenBank np 5713-9713, that includes 41% of total np whose nucleotide distribution is different between PD and controls, we found a statistically significant difference of lacunarity between PD and control groups with lower β values observed in the last one.
From literature, it is quite accepted that mitochondrial dysfunction and oxidative damage play an important role in the pathogenesis of PD as it is for other neurodegenerative diseases [32,33]. However, the mechanisms involved have not been clearly identified yet. Most mechanisms of mitochondrial dysfunction observed in PD are shared with the aging process [34,35] and overlapping of mtDNA alterations in PD and controls, without a clear-cut characterization of PD mtDNA mutation profile, observed also in this study when dealing with the whole mtDNA sequence, can explain the undefined genotype picture of this disease. As a matter of fact, although more deletions were found in mtDNA of PD patients, comparison with age-matched controls did not show any remarkable increase [35,36]. With the advent of more sensitive techniques to investigate the deleted mtDNA in individual cells, the increased number of mtDNA deletions in individual neurons of the substantia nigra has been confirmed in PD patients older than 65 years [36]. Another study [37], investigating the number of mtDNA deletions in the brains of subjects with parkinsonism, widespread Lewy body deposition, and less severe loss of dopaminergic neurons in substantia nigra, has reported a high proportion of mtDNA deletions (43%) in controls, which increased with age. However, a higher number of mtDNA deletions was observed in patients with parkinsonism and dementia (52%).
Generally, sequencing of mtDNA from PD patients has been performed in unselected groups, with and without a mitochondrial deficiency [38,39]. Although the results from some studies have suggested increased frequency of specific mtDNA polymorphisms in PD patients, this aspect has not been confirmed in all studies. In addition, PD has not been strongly associated with any specific mtDNA mutation yet. The increased frequency of specific mutations or mtDNA haplogroups observed in controls has suggested that they may play a protective role against PD, while some others have been associated with PD. Increased levels of somatic mtDNA point mutations in the substantia nigra have been found also in early PD, suggesting that such mutations occur in the early phase of this pathology [40-42]. If mtDNA mutations have any effect on this disease, their role appears to be complex and could involve specific haplotypes or combinations of sequence changes that modify mitochondrial function and make the system more vulnerable to nuclear genetic effects and/or environmental influences [43].
Such a picture highlights the usefulness of the theory of complexity and the laws of chaos to explain and characterize individual phenotypes evolving over time as ‘normal’ aging or pathological aging [1,6]. The inter-individual variability observed in senescent phenotype can be explained in the light of the theory of complexity by considering longevity as a “secondary product” of evolution of a dynamic nonlinear system. In fact, human beings as complex systems are made up of numerous sub-systems (nervous, endocrine, cardiovascular, … systems), interacting with each other. Each sub-system is further subdivided into interacting lower components (organs, tissues, cells, …) and so on at lower levels of organization. The organization into hierarchy and the laws of chaos can explain their evolution, from development to senescence, through the maintenance of a homeostatic dynamic equilibrium of their integrated functions as an adaptive response to continuous noxae from both endogenous and exogenous environments.
Based on this holistic point of view, aging has been defined as the temporal evolution of a complex system that evolves, under the influence of both endogenous and exogenous environments, with loss of complexity during aging [1,12,44]. Human beings, as complex systems characterized by a chaotic behavior, generate fractals that can be observed at both structural and functional levels. Fractal analysis, therefore, represents an intriguing tool useful to measure biocomplexity changes with aging and pathology. Fractal analysis can measure variations of complexity in biosystems that evolve with time by following different trajectories. The specific individual genetic-environment interactions determine the senescent phenotype that evolves as ‘normal’ aging, pathological aging, or successful aging [1,17]. Dealing with mutations of mtDNA as gaps in the nucleotide sequence, fractal lacunarity represents the most suitable tool to differentiate between PD and aging. In fact, lacunarity gives a holistic estimate of changes that occur in mtDNA sequences, comprising of number, type, and dislocation of mutation(s), the combination of which could contribute to PD onset and progression.
It is worth noting that a full certain diagnosis of PD is impossible during life: 75%–95% of PD patients have their diagnosis confirmed only post-mortem [45]. Diagnostic accuracy varies considerably depending on disease duration (lower on first visit), age, clinician expertise, and increasing understanding of this pathology. In this context, advances in live imaging, such as neuromolecular imaging, could improve the understanding as well as diagnosis of PD [46]. Often, failure in recognizing other pathologies causing neurodegenerative disorder or secondary parkinsonism as well as the absence of a true progressive parkinsonian disorder are common causes of diagnostic error [45].
Dealing with this and other observations discussed above, it would be interesting to verify the potential of our original method for mtDNA mutation-based profiling of neurodegenerative disease(s) also for the whole mtDNA sequence from PD patients as per AD [1]. In fact, we do not know whether the positive results observed on the smaller mtDNA frame but not on the whole sequence is peculiar to the disease or, rather, it is attributable to the sample of subjects used in this study. As a matter of fact, PD patients were diagnosed for late-onset PD; therefore, accumulation of mtDNA somatic mutations that occur with aging could mask PD specific point mutations or sequence changes of mtDNA. In addition, taking into account that PD is a neurodegenerative disease characterized by motor dysfunction, it would be useful to compare PD with age-matched controls without altered motor function. Our sample didn’t show any statistically significant difference as far as cognitive and motor function, unrelated to PD dysfunction, are concerned, the only difference accounting for limited head rotation in PD group. Last but not least characteristic of the sample under study deals with hospitalization: both PD patients and age-matched controls were hospitalized for other and different pathologies, probably with or without implication for mitochondrial dysfunction. In this context, it would be interesting to investigate whether the similar mtDNA mutation profile, observed in the whole mtDNA sequence of both PD and controls, could represent a mitochondrial genome profile peculiar to pathological senescent phenotype, characterized by multimorbidity affecting more than 60% elderly people that experience a pathological aging.
- Zaia, A.; Maponi, P.; Di Stefano, G.; Casoli, T. Biocomplexity and fractality in the search of biomarkers of aging and pathology: Focus on mitochondrial DNA and Alzheimer’s disease. Aging Dis. 2017, 8, 44–56, doi:10.14336/AD.2016.0629.
- Jeffrey, H.J. Chaos game representation of gene structure. Nucleic Acids Res. 1990, 18, 2163–2170, doi:10.1093/nar/18.8.2163.
- Zaia, A.; Eleonori, R.; Maponi, P.; Rossi, R.; Murri, R. Medical imaging and osteoporosis: Fractal’s lacunarity analysis of trabecular bone in MR images. In Proceedings of Eighteenth IEEE Symposium on Computer-Based Medical Systems—CBMS 2005, Dublin, Ireland, 23–24 June 2005; pp. 3–8, doi:10.1109/CBMS.2005.73.
- Zaia, A.; Eleonori, R.; Maponi, P.; Rossi, R.; Murri, R. MR imaging and osteoporosis: Fractal lacunarity analysis of trabecular Bone. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 484–489, doi:1109/TITB.2006.872078.
- Zaia, A.; Rossi, R.; Egidi, N.; Maponi, P. Fractal’s lacunarity analysis of trabecular bone in MR images. In Computational Vision and Medical Image Processing; Tavares, J., Jorge, N., Eds.; CRC Press: Boca Raton, FL, USA, 2010; pp. 421–426.
- Zaia, A. Fractal lacunarity of trabecular bone and magnetic resonance imaging: New perspectives for osteoporotic fracture risk assessment. World J. Orthop. 2015, 6, 221–235, doi:10.5312/wjo.v6.i2.221.
- de Rijk, M.C.; Breteler, M.M.; Graveland, G.A.; Ott, A.; Grobbee, D.E.; van der Meche, F.G.A.; Hofman, A. Prevalence of Parkinson’s disease in the elderly: The rotterdam study. Neurology 1995, 45, 2143–2146, doi:10.1212/WNL.45.12.2143.
- Huang, Z.; de la Fuente-Fernández, R.; Stoessl, A.J. Etiology of Parkinson’s disease. Can. J. Neurol. Sci. 2003, 30, S10–S18, doi:10.1017/S031716710000319X.
- Schapira, A.H.; Cooper, J.M.; Dexter, D.; Clark, J.B.; Jenner, P.; Marsden, C.D. Mitochondrial complex I deficiency in Parkinson’s disease. J. Neurochem. 1990, 54, 823–827, doi:10.1111/j.1471-4159.1990.tb02325.x.
- Hsieh, C.H.; Shaltouki, A.; Gonzalez, A.E.; Bettencourt da Cruz, A.; Burbulla, L.F.; St Lawrence, E.; Schüle, B.; Krainc, D.; Palmer, T.D.; Wang, X. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem. Cell 2016, 19, 709–724, doi:10.1016/j.stem.2016.08.002.
- Lill, C. M. (2016). Genetics of Parkinson's disease. Mol. Cell. Probes. 30, 386-396, doi:10.1016/j.mcp.2016.11.001.
- Andalib, S.; Vafaee, M.S.; Gjedde, A. Parkinson’s disease and mitochondrial gene variations: A review. J. Neurol. Sci. 2014, 346, 11–19, doi:10.1016/j.jns.2014.07.067.
- Simon, D.K.; Matott, J.C.; Espinosa, J.; Abraham, N.A. Mitochondrial DNA mutations in Parkinson’s disease brain. Acta Neuropathol. Commun. 2017, 5, 33, doi:10.1186/s40478-017-0433-9.
- Duarte, J.M.N.; Schuck, P.F.; Wenk, G.L.; Ferreira, G.C. Metabolic disturbances in diseases with neurological involvement. Aging Dis. 2014, 5, 238–255, doi:10.14336/AD.2014.0500238.
- Esteves, A.R.; Domingues, A.F.; Ferreira, I.L.; Januário, C.; Swerdlow, R.H.; Oliveira, C.R.; Cardoso, S.M. Mitochondrial function in Parkinson’s disease cybrids containing an nt2 neuron-like nuclear background. Mitochondrion 2008, 8, 219–228, doi:10.1016/j.mito.2008.03.004.
- Pettersson, M. Complexity and Evolution; Cambridge University Press: Cambridge, UK, 1996.
- Piantanelli, L.; Rossolini, G.; Basso, A.; Piantanelli, A.; Malavolta, M.; Zaia, A. Use of mathematical models of survivorship in the study of biomarkers of aging: The role of heterogeneity. Ageing Dev. 2001, 122, 1461–1475, doi:10.1016/s0047-374(01)00271-8.
- Zaia, A. Osteoporosis and fracture risk: New perspectives for early diagnosis and treatment assessment. In Osteoporosis: Etiology, Diagnosis and Treatment; Mattingly, B.E., Pillare, A.C., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2009; pp. 267–290.
- Grassberger, A.; Procaccia, I. Measuring the strangeness of strange attractors. Physica D 1983, 9, 189–208, doi:10.1016/0167-2789(83)90298-1.
- Goldberger, A.L.; Rigney, D.R.; West, B.J. Chaos and fractals in human physiology. Am. 1990, 262, 42–49, doi:10.1038/scientificamerican0290-42.
- Goldberger, A.L. Non-linear dynamics for clinicians: Chaos theory, fractals, and complexity at the bedside. Lancet 1996, 347, 1312–1314, doi:10.1016/s0140-6736(96)90948-4.
- Goldberger, L.A.; Peng, C.K.; Lipsitz, L.A. What is physiologic complexity and how does it change with aging and disease? Aging 2002, 23, 23–26, doi:10.1016/S0197-4580(01)00266-4.
- Mandelbrot, B.B. A Fractal’s Lacunarity, and how it can be Tuned and Measured. In Fractals in Biology and Medicine; Nonnenmacher, T.F., Losa, G.A., Weibel, E.R., Eds.; Birkhauser Press: Basel, Switzerland, 1993; pp. 8–21.
- Oiwa, N.N.; Glazier, J.A. Self-similar mitochondrial DNA. Biochem. Biophys.