Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Ali Movahedi and Version 2 by Sirius Huang.
Drought, the most significant environmental stressor, severely limits plant growth and development and significantly reduces crop production. Drought stress responses vary among plants, allowing them to withstand and survive adverse conditions. Plants resist drought by maintaining signaling pathways, such as the abscisic acid pathway, and activating unusual proteins, such as dehydrins.
- drought stress
- responsive proteins
- responsive pathways
- development
1. Introduction
To activate drought resistance mechanisms, plant cells must first be subjected to an imbalance in the water supply above and below ground and water loss. Several primary and secondary plant signaling pathways respond to drought stress by transmitting stress messages across gene networks [1][18]. A study on chickpea roots revealed that the gene network analysis involves Ca2+ signaling, chromatin organization, signal transduction, and interactions between proteins and their transcription (SuperSAGE) [2][19]. These networks include hormone signals, proteins, and metabolic compounds such as Reactive Oxygen Species (ROS), which are frequently required for gene expression. These compounds could be developed to stop cellular genomic damage [1][18]. Calcium signaling, ROS, Mitogen-Activated Protein Kinase (MAPK), phosphorylation cascades, and crosstalk between various TFs are all thought to be involved in stress perception and the stimulation of resistance and acclimation pathways [3][4][8,20]. Some drought-resistant enzymes have been found in the protein profiles of maize leaves, which are especially sensitive to drought. These enzymes are involved in the lignification, glycolysis, and Krebs cycles [5][21].
2. Calcium Signaling Pathways
Researchers have discovered that drought triggers a short-lived increase in the cytosolic Ca2+ concentration [2][19]. The role of Ca2+ in the response of crops to various environmental stimuli, such as drought stress, has long been recognized. Calmodulin (CaM), Calcineurin B-like (CBL), and Ca2+-Dependent Protein Kinases (CDPKs or CPKs) are three major calcium sensor families found in plants. CBLs are a distinct class of Ca2+ sensors found in plants that decode Ca2+ signals by triggering a group of plant-specific protein kinases identified as CBL-Interacting Protein Kinases (CIPKs) (Figure 1). CBLs are cytoplasmic Calcium-Binding Proteins (CaBs) that, by interacting with calmodulin, activate several protein kinases and phosphatases, thereby modulating downstream signaling [6][7][8][22,23,24]. CBL1, CBL4, and CBL9 are three CBL proteins that are present in most N-myristoylation patterns. These proteins play critical roles in abiotic stress signaling pathways in plants [7][9][10][23,25,26]. It was found that plants overexpressing CBL1 were better able to withstand drought [2][19]. Compared to CaM and CBL, CDPKs are the most frequently studied calcium sensors and have a unique structure that allows them to transmit Ca2+ signals via phosphorylation [7][11][23,27]. They have four conserved calcium-binding motifs, a variable domain, an auto-inhibitory connector region, a Ser/Threonine protein kinase domain (Ser/Thr), and a regulatory Calmodulin-Like Domain (CaM-LD). A CaM-LD with a Ser/Thr kinase domain is another component of CDPKs. CDPKs contain four known components that assist plants in detecting environmental stimuli, transferring calcium signals, and mediating various cellular responses [2][9][11][19,25,27].
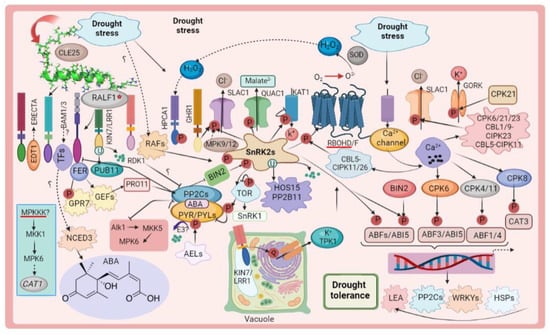
Figure 1. The PYR/Pyls/RCAR (clade A type 2C protein phosphatase receptors) are capable of recognizing ABA. The anion channels SLAC1 and QUAC1 are activated and phosphorylated by phosphorylated OST1. Calcium-dependent protein kinases can decode a particular Ca2+ signature under drought stress. During stress, membrane-anchored receptor-like kinases detect extracellular signals and transfer signals within cells. An interaction between RALF1, FER, and the GEF, ROP11, is required for ABI2 activation. Various protein kinases, such as CPKs and CIPKs, RLKs, SnRK2s, and MAPKs, work together to coordinate plant responses to drought stress. Positive and negative regulations are indicated by arrows and bars, respectively. Solid lines represent direct regulation, while dashed lines or unknown regulations represent indirect regulation.
3. Phytohormone-Mediated Pathways
The phytohormone ABA plays a key role in the complex adaptations made by plants in response to drought stress, including the closure of stomata and the activation of drought-responsive genes [12][28]. ABA is essential for generating hormones and osmotic signals in response to drought. This reaction conceals three primary drought-coping mechanisms: the ability to flee dry conditions, to avoid becoming dehydrated, and to tolerate dryness or desiccation [12][13][14][6,28,29]. It has been shown that chloroplast enzymes such Zeaxanthin Epoxidase (ZEP), Nine-Cis-Epoxycarotenoid Dioxygenase (NCED), and Abscisic Aldehyde Oxidase (AAO) contribute to the biosynthesis of ABA in plants [15][30]. Plants respond to drought through ABA-dependent and ABA-independent signaling pathways [16][31]. It is well established that NAC TFs are responsible for controlling the expression of genes that participate in the ABA pathway, Pyrabactin Resistance (PYR) and Pyrabactin Resistance 1-Like (PYL), and the induction of the production of the ABA hormone. Overexpression of the stress-responsive gene SNAC1 (STRESS-RESPONSIVE NAC 1) was shown to dramatically enhance drought resistance in transgenic rice (22–34% more) in a research area under intense drought stress at the reproductive phase without causing any phenotypic changes or yield penalty. Drought mainly causes the transcription factors NAM, ATAF, and CUC (NAC), which have transactivation activity, to be activated in guard cells [17][32]. OsNAP, a transcription factor that is nucleus-localized and a member of the NAC family, functions as a transcriptional activator in yeast. This gene was shown to be highly activated in rice by ABA and abiotic stressors, such as drought and low temperature, according to an analysis of OsNAP transcription levels. Rice plants that overexpressed OsNAP were not found to exhibit growth delays, but they did exhibit a considerably decreased rate of water loss, greater tolerance to high salt, drought, and low temperature conditions during the vegetative stage and a higher yield under drought stress during the flowering stage [18][33].
4. MAPK-Dependent Pathways
The MAPKs are a family of proteins with a widespread distribution and substantial evolutionary conservation. Plant MAPK cascades control a wide range of developmental and stress-response functions. In response to various abiotic and biotic stresses, including drought, saltiness, high and low temperatures, injury, and pathogenic incursion, MAPKs are involved in several critical cellular processes [19][34]. Signaling cascades rely heavily on transiently phosphorylated MAPKs, including MAPK Kinases (MAPKKs) and MAPK Kinase Kinases (MAPKKKs). Multiple upstream receptors and different downstream target components are influenced by the MAPK, MAPKK, and MAPKKK network in distinct signal transduction pathways [20][35].
The MAPK family is activated consecutively via protein phosphorylation. An active MAPKKK stimulates an MAPKK through threonine phosphorylation during kinase activity, which stimulates an MAPK [21][36]. In response to different stress signals, activated MAPK phosphorylates cytoskeletal proteins and phospholipases, which leads to the expression of genes [22][37]. RNA-Seq research has shown that several members of the MAPK cascade change in response to drought in plants. In response to drought stress, rice produces transcripts of OsMPK4, OsMKK1, OsMKK4, OsMPK5, OsMPK7, and OsMPK8 [23][38]. In wheat, the levels of expression of TaRaf44, TaMPK8, TaRaf72, TaRaf87, TaMKK1, TaRaf105, TaRaf80, and TaMKKK16 were shown to be altered after drought stress [24][39]. The transcription levels of cotton GhMPK gene families, such as 2, 4, 6, 7, 17, and 31, were found to be drastically reduced under drought stress conditions, whereas cotton GhMEKK gene families, such as 10, 12, 24, and 36 and GhRAF4, were shown to be increased following exposure to drought stress [24][39]. In addition, dry circumstances in maize triggered the expression of several drought-inducible genes, including ZmMPK3 and ZmMPK15, and the ZmMKK10-2 and ZmMAPKKK gene families [24][39]. These results show the significance of MAPKs in drought, although our understanding of their biological roles during drought stress is still unclear.
