Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Athanasios Armakolas.
Circulating tumor cells (CTCs) are a population of cancer cells that manage to detach from either the primary tumor or metastatic deposits in the periphery of patients, and they seem to have a short half-life of approximately 1h to 2.4 h.
- cancer
- diagnosis
- prognosis
- CTCs
- ctDNA
1. Introduction
Metastasis is a multi-step process that depends on the presence of CTCs in the blood stream and/or disseminated tumor cells (DTCs) that are found in the bone marrow [7,8][1][2]. In order for CTCs to be able to disseminate from primary tumors they must undergo phenotypic changes that will allow the cells to penetrate blood vessels [10][3]. The epithelial-mesenchymal transition (EMT) is a central process in metastasis where the cancer epithelial cells downregulate the expression of their epithelial markers, including the cell membrane proteins that are responsible for cell to cell adhesion, and they express the mesenchymal markers [11,12,13][4][5][6]. Mesenchymal cells do not possess cell adhesion molecules on their surface and, therefore, can easily detach from the main tumor. In addition these cells induce the expression of proteases and integrins that are central molecules in the intravasation and extravasation of mesenchymal cancer cells [14,15][7][8]. CTCs are a population of cancer cells that manage to detach from either the primary tumor or metastatic deposits in the periphery of patients, and they seem to have a short half-life of approximately 1h to 2.4 h. The presence of CTCs in the bloodstream consists of a very heterogenous population that varies greatly in number from patient to patient and even within the same patient at different time points.
The presence of CTCs in the circulation is fundamental for the development of metastasis in various types of solid tumors [16,17][9][10] (Figure 1). The fact that CTCs are highly heterogeneous and circulate in low numbers renders them a very hard target to detect accurately enough to set the guidelines for patient treatment (Figure 1). Therefore, it can easily be understood that the first step of enrichment is critical for the analysis of the cancer cell load (metastatic or not) in the periphery of the patients. For this reason, a variety of techniques have been developed, based on both the biophysical and biological properties of these cells, in order to differentiate them from their background and enrich them, so that they are compatible with molecular analysis or imaging analysis. Similarly, many technologies have been developed for the capture, isolation and detection of CTCs [18,19,20,21,22][11][12][13][14][15].
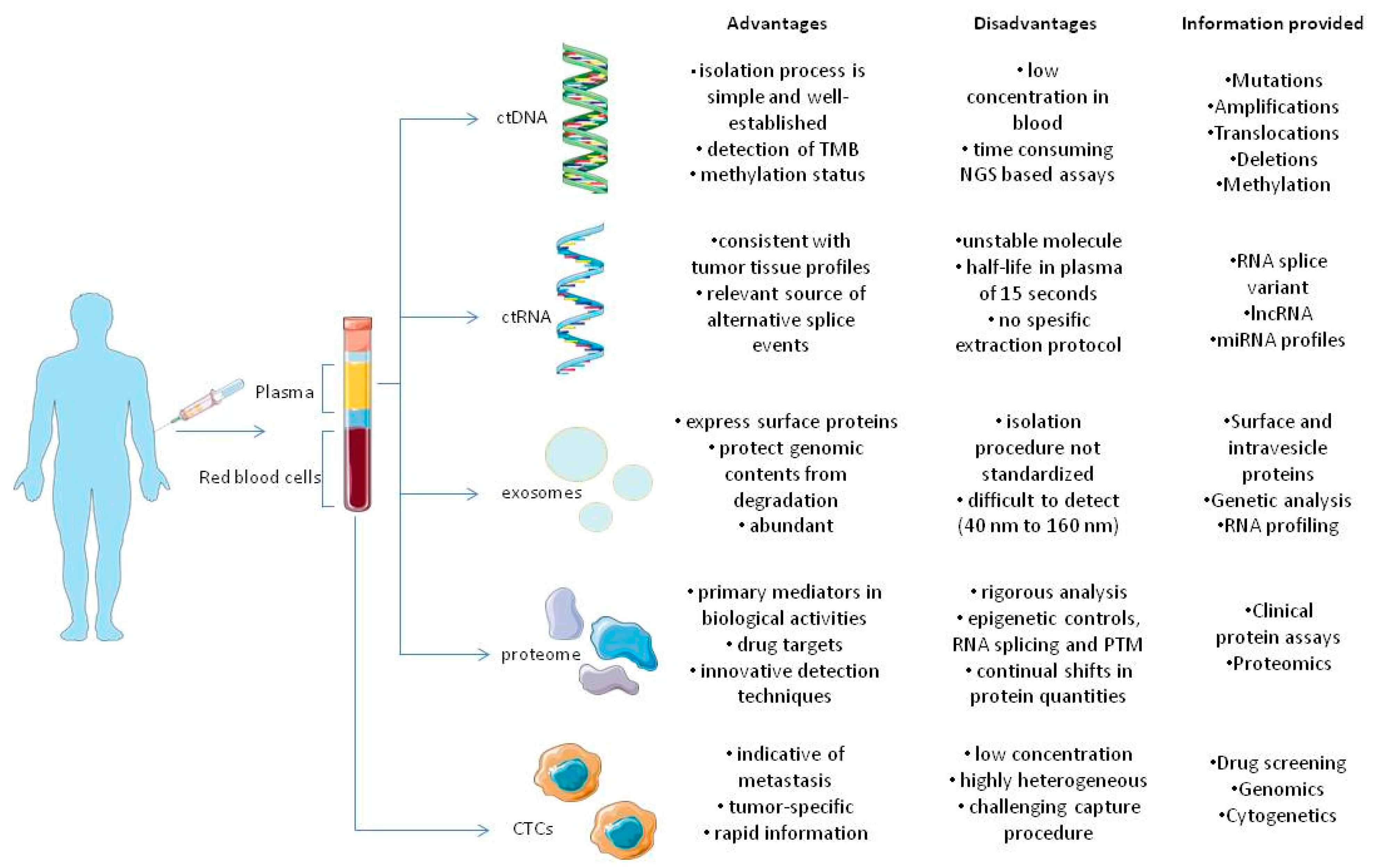
Figure 1. Components of tumor circulome examined in liquid biopsies and their applications. The several analytes extracted from blood provide a wide variety of information regarding tumors. As previously stated, all analytes share different advantages and disadvantages that favor or oppose their usage in clinical settings, in tumor diagnosis, monitoring and therapy [23][16].
2. Enrichment and Isolation of CTCs
Due to the small CTC concentration in the blood, analysis always starts with an enrichment step that aims to increase the concentration of these cells by several logarithmic units thus allowing an easier identification of single tumor cells. CTCs can be enriched by approaches that exploit differences between tumor and normal blood cells, based on biological properties such as the differential expression of protein markers or different physical properties of cells including size, density, deformability or electric charges, and these enrichment principles can be combined to optimize the yield of CTCs [17,24,25][10][17][18].
A variety of devices has been developed to enrich and detect CTCs, with emphasis on devices capable of selecting and detecting CTCs that have undergone EMT [25,26][18][19]. The EPCAM-based enrichment for CTC detection has provided a reliable prognostic tool in different carcinomas [27,28][20][21]. However other epithelial cell surface antigens including EGFR7 [29][22] and mucin 1 [30][23], and tissue specific antigens such as prostate specific membrane antigen (PSMA) [31][24] for prostate cancer cells and ERBB2 [32][25] for breast cancer cells have been exploited for this purpose (Table 1). CTCs present a very heterogeneous population of cells. Recent evidence indicates that in many cases CTCs could cease to express the selected marker, leading to markers escaping detection and, thus, to false negative results [33,34][26][27]. Consequently, the bias which might be introduced by positive selection can be avoided by negative selection. In this case non-malignant blood cells are depleted from the blood using antibodies that recognize the cell surface antigens expressed on leukocytes, usually CD45 and other cells in the bloodstream, including endothelial stem cells with markers such as CD146 and hematopoietic stem cells with markers such as CD34 [35,36][28][29]. Disadvantages of negative selection include the lower purity of isolated CTC populations compared to the techniques of positive selection and the risk of CTCs becoming trapped in a mass of blood cells and, thus, being included in the depleted cell fraction and ignored [37,38,39][30][31][32].
With regard to techniques based on the physical differences of tumor cells and non-malignant blood cells, it is worth noting that these characteristics are highly variable between CTCs and have substantial overlap with those of non-malignant cells; therefore, definitions of CTC size depend on the capture device. Microfiltration technologies have been developed where blood is passed through pores or microfluidic passageways with calibrated size to trap CTCs, resulting in size exclusion and, therefore, retention of large CTCs, albeit with possible loss of small CTCs [40][33]. Other microfluidic devices that rely on size separation use inertial focusing strategies to separate CTCs from other blood components, while dielectrophoresis (DEP) allows the separation of CTCs based on the different electrical charges of tumor and blood cells. Special microfiltration systems have also been developed to specifically capture CTC clusters based on size exclusion [41][34]. Most CTCs occur as single cells, but CTC clusters can be detected and their biology is still being investigated [42][35].
Table 1.
Clinical applications of CTCs.
| Marker | Assay Relevance | Disease | Technique | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|
| ERBB2 | Prognostic/guiding therapy | Breast cancer | CellSearch | First-line ERBB2-targeted therapy for metastatic breast cancer appears to reduce CTC levels more than endocrine or chemotherapeutic therapy; anti-ERBB2 therapy appears to be linked with lower total CTC levels. | Retrospective analysis. The small number of patients with progressive disease highlights the implicit difficulties in analyzing the rate of CTC-positive cases. |
[43][36] |
| PDL-1 | Guiding therapy | Breast cancer | CellSearch | CTC and platelet PD-L1 expression might be used to determine which patients should be treated with immune checkpoint inhibitors and as a pharmacodynamic biomarker during therapy. | The CellSearch platform might potentially disrupt CTC doublets and clusters, underestimating their biological/prognostic effect compared with other assays. | [44][37] |
| EpCAM | Prognostic | Metastatic breast cancer | CellSearch | CTC counts as independent prognostic factor. CTC testing is more repeatable than radiology and detects progression earlier. |
The findings of the study do not support the use of the test as a screening tool for detecting new primary or metastatic breast cancer. | [45,46,47][38][39][40] |
| Nonmetastatic primary breast cancer | Laser scanning cytometry | A rise in CTC counts of more than ten-fold at the conclusion of therapy is significantly predictive of recurrence. | The relationship among CTC number and therapeutic efficacy may change amongst patients. | [48][41] | ||
| CellSearch | CTCs are considered an independent prognostic factor. | CTC detected only in 24% of patients studied. | [49,50][42][43] | |||
| Non-small-cell lung cancer | CellSearch | CTC reduction as an early indicator of therapeutic response. | Biomarkers’ predictive or prognostic value cannot be differentiated. | [51][44] | ||
| Small-cell lung cancer | CellSearch and ISET | CTC isolation using ISET is dependent on cellular size and independent of any cellular marker. | NA | [52][45] | ||
| Metastatic colorectal cancer | CellSearch | CTC count as independent prognostic factor. | Overall CTC yield is less than in other epithelial malignancies such as breast cancer. | [53][46] | ||
| Non metastatic colorectal cancer | CellSearch | CTC count as independent prognostic factor. | It is uncertain if CTCs discovered are precursors of metastatic lesions or if CTCs arise from metastases and are just a marker of overall disease burden. | [54][47] | ||
| Hepatocellural cancer | CellSearch and EpCAM-based immunoenrichment and FACS | The study demonstrated the feasibility of utilizing CTC-derived DNA for next-generation sequencing. | NA | [55][48] | ||
| Castration-resistant prostate cancer | CellSearch | At all time points, CTC numbers predicted OS better than PSA decrement methods. | NA | [56,57][49][50] | ||
| ALK | Predictive | Lung adenocarcinoma | ISET-ICC/FISH | Noninvasive real-time monitoring of targeted treatment is a possibility. | NA | [58][51] |
| ASGR1 | Prognostic | Hepatocellular carcinoma | Antibody-coated magnetic beads based separation | In terms of specificity and sensitivity, CTC detection exceeds AFP mRNA. | Comparison with other studies is problematic owing to the unique technique utilized. | [59][52] |
| PSA, PSMA, PSCA, KRT19 | Prognostic | Prostate cancer | RT-PCR | High sensitivity of RT-PCR. | False positive and false negative results are possible. | [60][53] |
| Pan-cytokeratin, AR-V7, CD45 | Prognostic/predictive | Metastatic castration-resistant prostate carcinoma | AdnaTest | Taxanes as chemotherapeutic drugs of choice for individuals with androgen receptor signaling blocker resistance (based on CTC AR-V7 positivity). | Due to the small sample size, multivariable analysis to connect AR-V7 status with prognosis and define subpopulations was not possible. | [61][54] |
| Epic AR-V7 Test | Stratification for CTH based on taxanes in mCRPC CTC AR-V7+. | NA | [62,63][55][56] | |||
| Tyrosinase | Prognostic | Malignant melanoma | RT-PCR | CTC count as independent prognostic factor. | NA | [64][57] |
| MART1, MAGE-A3, and PAX3 | Prognostic | Malignant melanoma | RT-PCR | CTC count as independent prognostic factor. | The presence of CTCs was not related to any of the identified clinical prognostic indicators. | [65][58] |
3. Detection of CTCs
After enrichment, an identification step is required to detect CTCs surrounded by residual leukocytes at the single cell level by immunological, molecular or functional methods [66][59]. The dominant methods use antibodies against membrane and cytoplasmic antigens, including epithelial, mesenchymal, histospecific and tumor-related markers, with the aim of direct immunological detection [26][19]. Until now, the only clinical application of CTCs approved by the FDA is the CellSearch platform [28][21] and the most current CTC assays use the same identification step as this one (Table 1). Cells stained with fluorescently labeled antibodies to epithelial cytokeratin (CK) are visualized through fluorescence microscopy and used as a marker of CTCs, while staining of CD45 is used to exclude leukocytes [67][60]. Some of the markers used vary in different types of cancer, for example cytokeratins apply to breast, colon and prostate cancers, and other epithelial tumors, although specific tissue antigens can also be used, such as prostate specific antigen (PSA) or breast specific mammaglobin [17][10]. However, this technology has some limitations. First of all, it is mainly based on the expression of EpCAM which has been associated with localized cancer, but during metastasis its expression, along with that of CK, decreases amid the appearance of mesenchymal markers [68][61]. Second, cell isolation through the CellSearch system is followed by cell fixation for stabilization, which prevents further characterization of viable cells such as CTC cultures, while having a low sensitivity for CTC detection (one cell per 1 mL of blood sample). Finally, the CellSearch system offers low purity of captured cells, in the range of 60–70%, resulting in captured CTCs that are usually contaminated by blood cells or normally circulating epithelial cells (CECs) [42,65,69][35][58][62].
It is possible to manually isolate the identified CTCs by micromanipulation; however, it is laborious and time-consuming. An alternative approach for separation of CTCs in order to further genomic, molecular or functional analyses involves automated selection of single cells using a DEPArray, a device that allows trapping single CTCs in DEPcages [70][63]. Dielectrophoresis (DEP) is a liquid biopsy separation assay that is based on particles with different polarizations and that move differently under a non-uniform electric field [71][64]. Microchips that use the DEP method have multiple integrated electrodes which generate the non-uniform electric field in order to isolate and capture single CTCs. However, the low sample volumes and the varying dielectric features of cells due to ion leakage could limit the isolation time [72][65].
A new fluorescence-activated cell sorting (FACS) approach has also been used for CTC detection and phenotypic analysis, but this technology typically requires a pre-enrichment step to achieve sufficiently high initial CTC concentrations [73][66]. FACS is a cell-based analytic method where an immunomagnetically enriched blood sample is injected into a fluid stream, and single cells in the stream are interrogated by lasers as they flow into a capillary tube. The cells are then sorted based on light scattering and fluorescence patterns by comparison with negative (healthy blood cells) and positive control (EpCAM-expressing cancer cell lines) [74][67]. On the other hand, there are some restrictions to this method. Τhe use of expensive antibodies leads to high detection costs, whereas, in many cases CTCs cannot be further analyzed in real-time conditions since the cells are fixed or lysed during the assay process [75,76][68][69].
CTCs can also be detected by techniques that target the mRNA or DNA level. These techniques require the design of PCR tests with specific primers for tissue, organ, tumor-specific transcriptions, or for tumor-specific mutations, translocations or methylation patterns unique to the tumor [77][70]. Furthermore, these technologies also allow the quantification of CTC numbers. Reverse transcription PCR (RT-PCR) assays are the most user-friendly method for detecting low-abundance mRNA transcriptions. A limitation of this approach is that the CTC number can only be estimated due to the fact that gene expression levels vary between CTCs [78][71]. Currently, digital droplet PCR (ddPCR) allows detection and absolute quantification of low-abundance targets in shorter times without requiring a large number of replications [79,80][72][73]. ddPCR relies on water-oil emulsion droplet technology. In comparison with other digital PCR assays, this method has a lower sample requirement, thereby reducing costs and preserving valuable samples [81][74].
Functional assays like the epithelial ImmunoSPOT (EPISPOT) assay have been used for CTC in vitro detection in blood and bone marrow samples for more than two decades and have been validated at the clinical level for several different cancers [82][75]. This assay provides quantitative information about the number of viable CTCs present in the sample based on the fluorescence detection of specific epithelial proteins secreted by these cells, as well as qualitative information about which of these proteins are shed during cell culture. Currently, this technique has been further developed allowing for the capture and detection of CTCs at the single cell level. The so-called EPIDROP, as an abbreviation of ELISPOT in a drop, is a more rapid and sensitive form than the previous one [83][76]. In this assay, CTCs are immunostained prior to individual encapsulation in fluid microdroplets and, consequently, both the total number of CTCs (EPCAM+ or EPCAM−) and the number of functional CTCs can be imprinted. Indeed, viable CTCs can be distinguished from apoptotic CTCs, and EPCAM+ versus EPCAM− CTCs enable the assessment of EMT status. In the future, a subsequent molecular characterization of the captured CTCs will be incorporated into this innovative assay. Despite the fact that EPISPOT is a promising technique, there can be problems when antigen levels are lower or binding efficiency is reduced [84][77]. Furthermore, processing a single sample in an EPISPOT assay requires three days for analysis [85][78]. This, in combination with the finding that it may fail to isolate more heterogeneous cells because of its biomarker dependence [86][79], render EPISPOT unsuitable for clinical use.
4. Characterization of CTCs and Analytical Technologies
Aiming towards personalized cancer treatment, many innovative technologies have been developed in recent years intended for the characterization of CTCs. CTCs can be analyzed by cytogenetic analyses, such as in situ fluorescence hybridization (FISH), to identify chromosomal rearrangements [87][80] (Table 1). Multi-omics techniques have entered dynamically in the patient management of cancer. CTC single cell analysis is a novel approach where CTCs are isolated and the entire genome can be amplified in order to make subsequent assessments of duplicate number aberrations and specific mutations using array competitive genome hybridization or next-generation sequencing (NGS) techniques [88][81]. A physical disadvantage of this method is that the findings cannot be verified because single CTCs are found in limited quantities [89][82]. Though, the DNA amplification protocol requires careful technical validation to avoid false findings and, thus, ensures a low error rate [62][55]. In addition, strict bioinformatic approaches are needed to ensure reliable identification of tumor-specific changes in individuals’ CTCs. Therefore, it is a time-consuming technique that also involves high costs [90][83] (Figure 1). On the other hand, this method offers high sensitivity of CTCs from many tumor types, and the variety of selection markers allows for the possibility of characterizing cells for multiple markers all at the same time [91][84]. In addition, the use of NGS in CTC analysis offers the possibility of using genomic and transcriptional CTC profiles to improve the understanding of cancer heterogeneity [92][85].
RT-PCR transcription assays that are not expressed in non-malignant blood cells such as those encoding PSA or epithelial cytokeratins are sensitive enough to allow the detection of single CTCs but can also provide information on their phenotype (Table 1). However, special manipulations are required because low-level external expression of target transcript in infected leukocytes (or other non-malignant cells in the bloodstream) can lead to incorrect attribution of the results (false positives). As an alternative approach, the transcriptomic profile of single CTCs isolated by micromanipulation can be determined using multiplex quantitative RT-PCR [93][86] or RNA sequencing assays, NGS. These techniques may also allow the assessment of heterogeneity between single CTCs within the same patient [94][87]. The sensitivity of NGS-based technologies is lower than that of PCR-based technologies and inversely proportional to the number of sites analyzed, with the total exome sequence (WES) having the lowest sensitivity [95][88]. On the other hand, compared to ddPCR, NGS had a higher sensitivity for individual nucleotide variants, indels and selected rearrangements and has been shown to have a positive percentage agreement of 95% and a positive predictive value of 100% [96][89].
On the other hand, immunophenotyping with antibodies to proteins of interest (proliferation or apoptosis markers) is the most commonly used approach to CTC characterization but is currently limited to a few proteins of interest (beyond those required for the enrichment and detection). In many studies immunophenotyping has been used to confirm the epithelial [97][90] or mesenchymal [98][91] nature of the suspected circulating cells (Table 1). However, even among the epithelial markers typically used to conceive CTCs, such as EpCAM or cytokeratins, there is no consensus on specific markers that can more effectively identify clinically relevant CTCs.
A micro-fluid single cell western blot (scWB) technology has also been developed for proteomic CTC phenotyping but is limited to evaluating only eight proteins [99][92].The rare cell scWB quantifies multiple surface and intracellular signaling proteins in each individual CTC, allowing estimates of the variation in biological protein expression between CTCs. This method is compatible with well-established CTC isolation tools and can successfully analyze CTC populations with just two primary cells. The monitoring of multiple regulated proteins in blood derived CTC may provide information about the treatment options to maximize the benefit for each specific patient at each specific time point [100][93].
With in vitro cultures of CTCs, in addition to transient expansion, some groups have been able to create permanent CTC cell lines obtained from patients with advanced-stage diseases. However, these cell lines have phenotypes that reflect those of cells in tumor tissue samples from patient donors, but they also have a special molecular signature that reflects the metastasizing capacity generally attributed to CTCs. In practice, cell lines derived from CTCs have germinality, a specific DNA repair phenotype and a high metabolic rate [101][94].These cell lines can also be used to test drugs in prospective discovery projects, but the process of determining these cell lines is not yet fast enough and CTCs capable of metastasis are a rare subset of the cell population, thus limiting the usefulness of this approach for decision-making in clinical practice.
Furthermore, short-term CTC cultures could provide information quickly enough to potentially inform treatment decisions for the donor patient. They could also reveal new pathways specific to metastasis-causing CTCs and, therefore, new targets for drugs that specifically eliminate this more aggressive subset of CTCs. The evolution of CTCs presents another challenge for the development of cell lines that accurately reflect the disease, and the creation of multiple cell lines using CTCs isolated from sequential blood samples collected during disease and treatment can provide unique information [102][95].
Finally, CTCs can also be characterized through functional studies in patient-derived xenograft models (PDX) which can result in revealing the properties of these cells that are required for the transition to secondary sites and/or the outgrowth of diffuse cancer cells (DTCs) to form apparent metastases. In addition, these PDX models can be used to test drugs that may be interesting candidates for anticancer therapy [103][96] (Figure 1). The disadvantage of this method, however, is that the development of PDX models usually takes several months and the rate of successful CTC integration is generally very low due to the requirement of a large number of CTCs, which generally excludes the use of such models in making treatment decisions for individual patients. However, these models appear to recap the molecular and cellular characteristics of parent tumors as well as the response to chemotherapy [86,104][79][97].
References
- Dasgupta, A.; Lim, A.R.; Ghajar, C.M. Circulating and Disseminated Tumor Cells: Harbingers or Initiators of Metastasis? Mol. Oncol. 2017, 11, 40–61.
- Alix-Panabières, C. EPISPOT Assay: Detection of Viable DTCs/CTCs in Solid Tumor Patients. In Minimal Residual Disease and Circulating Tumor Cells in Breast Cancer; Recent Results in Cancer, Research; Ignatiadis, M., Sotiriou, C., Pantel, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 69–76. ISBN 978-3-642-28160-0.
- Lozar, T.; Gersak, K.; Cemazar, M.; Kuhar, C.G.; Jesenko, T. The Biology and Clinical Potential of Circulating Tumor Cells. Radiol. Oncol. 2019, 53, 131–147.
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773.
- Serrano-Gomez, S.J.; Maziveyi, M.; Alahari, S.K. Regulation of Epithelial-Mesenchymal Transition through Epigenetic and Post-Translational Modifications. Mol. Cancer 2016, 15, 18.
- Diaz, J.M.; Wagner, M.S.; Sousa-Squiavinato, A.C.; de Freitas-Junior, J.C.; de Araújo, W.M.; Tessmann, J.W.; Rocha, M.R. Epithelial-Mesenchymal Transition in Metastatic Colorectal Cancer. Exon. Publ. 2022, 25–42.
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335.
- Canel, M.; Serrels, A.; Frame, M.C.; Brunton, V.G. E-Cadherin-Integrin Crosstalk in Cancer Invasion and Metastasis. J. Cell. Sci. 2013, 126, 393–401.
- Di Capua, D.; Bracken-Clarke, D.; Ronan, K.; Baird, A.-M.; Finn, S. The Liquid Biopsy for Lung Cancer: State of the Art, Limitations and Future Developments. Cancers 2021, 13, 3923.
- Pantel, K.; Alix-Panabières, C. Liquid Biopsy and Minimal Residual Disease—Latest Advances and Implications for Cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424.
- Markou, A.; Tzanikou, E.; Lianidou, E. The Potential of Liquid Biopsy in the Management of Cancer Patients. Semin. Cancer Biol. 2022, 84, 69–79.
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating Tumor Cell Isolation, Culture, and Downstream Molecular Analysis. Biotechnol. Adv. 2018, 36, 1063–1078.
- Xiao, J.; Pohlmann, P.R.; Isaacs, C.; Weinberg, B.A.; He, A.R.; Schlegel, R.; Agarwal, S. Circulating Tumor Cells: Technologies and Their Clinical Potential in Cancer Metastasis. Biomedicines 2021, 9, 1111.
- Ju, S.; Chen, C.; Zhang, J.; Xu, L.; Zhang, X.; Li, Z.; Chen, Y.; Zhou, J.; Ji, F.; Wang, L. Detection of Circulating Tumor Cells: Opportunities and Challenges. Biomark. Res. 2022, 10, 58.
- Kowalik, A.; Kowalewska, M.; Góźdź, S. Current Approaches for Avoiding the Limitations of Circulating Tumor Cells Detection Methods—Implications for Diagnosis and Treatment of Patients with Solid Tumors. Transl. Res. 2017, 185, 58–84.e15.
- SMART. Available online: https://smart.servier.com/ (accessed on 18 January 2023).
- Deng, Z.; Wu, S.; Wang, Y.; Shi, D. Circulating Tumor Cell Isolation for Cancer Diagnosis and Prognosis. eBioMedicine 2022, 83, 104237.
- Gires, O.; Pan, M.; Schinke, H.; Canis, M.; Baeuerle, P.A. Expression and Function of Epithelial Cell Adhesion Molecule EpCAM: Where Are We after 40 Years? Cancer Metastasis Rev. 2020, 39, 969–987.
- Alix-Panabières, C.; Mader, S.; Pantel, K. Epithelial-Mesenchymal Plasticity in Circulating Tumor Cells. J. Mol. Med. 2017, 95, 133–142.
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836.
- Riethdorf, S.; O’Flaherty, L.; Hille, C.; Pantel, K. Clinical Applications of the CellSearch Platform in Cancer Patients. Adv. Drug Deliv. Rev. 2018, 125, 102–121.
- Ohnaga, T.; Takei, Y.; Nagata, T.; Shimada, Y. Highly Efficient Capture of Cancer Cells Expressing EGFR by Microfluidic Methods Based on Antigen-Antibody Association. Sci. Rep. 2018, 8, 12005.
- Thege, F.I.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Kochman, M.L.; Hollingsworth, M.A.; Rhim, A.D.; Kirby, B.J. Microfluidic Immunocapture of Circulating Pancreatic Cells Using Parallel EpCAM and MUC1 Capture: Characterization, Optimization and Downstream Analysis. Lab Chip 2014, 14, 1775–1784.
- El-Heliebi, A.; Hille, C.; Laxman, N.; Svedlund, J.; Haudum, C.; Ercan, E.; Kroneis, T.; Chen, S.; Smolle, M.; Rossmann, C.; et al. In Situ Detection and Quantification of AR-V7, AR-FL, PSA, and KRAS Point Mutations in Circulating Tumor Cells. Clin. Chem. 2018, 64, 536–546.
- Hyun, K.-A.; Lee, T.Y.; Lee, S.H.; Jung, H.-I. Two-Stage Microfluidic Chip for Selective Isolation of Circulating Tumor Cells (CTCs). Biosens. Bioelectron. 2015, 67, 86–92.
- Wu, L.-L.; Tang, M.; Zhang, Z.-L.; Qi, C.-B.; Hu, J.; Ma, X.-Y.; Pang, D.-W. Chip-Assisted Single-Cell Biomarker Profiling of Heterogeneous Circulating Tumor Cells Using Multifunctional Nanospheres. Anal. Chem. 2018, 90, 10518–10526.
- Paoletti, C.; Larios, J.M.; Muñiz, M.C.; Aung, K.; Cannell, E.M.; Darga, E.P.; Kidwell, K.M.; Thomas, D.G.; Tokudome, N.; Brown, M.E.; et al. Heterogeneous Estrogen Receptor Expression in Circulating Tumor Cells Suggests Diverse Mechanisms of Fulvestrant Resistance. Mol. Oncol. 2016, 10, 1078–1085.
- Rapanotti, M.C.; Campione, E.; Suarez Viguria, T.M.; Spallone, G.; Costanza, G.; Rossi, P.; Orlandi, A.; Valenti, P.; Bernardini, S.; Bianchi, L. Stem–Mesenchymal Signature Cell Genes Detected in Heterogeneous Circulating Melanoma Cells Correlate With Disease Stage in Melanoma Patients. Front. Mol. Biosci. 2020, 7, 92.
- Hsieh, J.C.-H.; Wu, T.M.-H.; Hsieh, J.C.-H.; Wu, T.M.-H. The Selection Strategy for Circulating Tumor Cells (CTCs) Isolation and Enumeration: Technical Features, Methods, and Clinical Applications; IntechOpen: London, UK, 2016; ISBN 978-953-51-2631-7.
- Habli, Z.; AlChamaa, W.; Saab, R.; Kadara, H.; Khraiche, M.L. Circulating Tumor Cell Detection Technologies and Clinical Utility: Challenges and Opportunities. Cancers 2020, 12, 1930.
- van der Toom, E.E.; Verdone, J.E.; Gorin, M.A.; Pienta, K.J. Technical Challenges in the Isolation and Analysis of Circulating Tumor Cells. Oncotarget 2016, 7, 62754–62766.
- Chu, P.-Y.; Hsieh, C.-H.; Wu, M.-H. The Combination of Immunomagnetic Bead-Based Cell Isolation and Optically Induced Dielectrophoresis (ODEP)-Based Microfluidic Device for the Negative Selection-Based Isolation of Circulating Tumor Cells (CTCs). Front. Bioeng. Biotechnol. 2020, 8, 921.
- Sun, N.; Li, X.; Wang, Z.; Li, Y.; Pei, R. High-Purity Capture of CTCs Based on Micro-Beads Enhanced Isolation by Size of Epithelial Tumor Cells (ISET) Method. Biosens. Bioelectron. 2018, 102, 157–163.
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A Microfluidic Device for Label-Free, Physical Capture of Circulating Tumor Cell Clusters. Nat. Methods 2015, 12, 685–691.
- Gkountela, S.; Castro-Giner, F.; Szczerba, B.M.; Vetter, M.; Landin, J.; Scherrer, R.; Krol, I.; Scheidmann, M.C.; Beisel, C.; Stirnimann, C.U.; et al. Circulating Tumor Cell Clustering Shapes DNA Methylation to Enable Metastasis Seeding. Cell 2019, 176, 98–112.e14.
- Deutsch, T.M.; Riethdorf, S.; Fremd, C.; Feisst, M.; Nees, J.; Fischer, C.; Hartkopf, A.D.; Pantel, K.; Trumpp, A.; Schütz, F.; et al. HER2-Targeted Therapy Influences CTC Status in Metastatic Breast Cancer. Breast Cancer Res. Treat. 2020, 182, 127–136.
- Paoletti, C.; Miao, J.; Dolce, E.M.; Darga, E.P.; Repollet, M.I.; Doyle, G.V.; Gralow, J.R.; Hortobagyi, G.N.; Smerage, J.B.; Barlow, W.E.; et al. Circulating Tumor Cell Clusters in Patients with Metastatic Breast Cancer: A SWOG S0500 Translational Medicine Study. Clin. Cancer Res. 2019, 25, 6089–6097.
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. New Engl. J. Med. 2004, 351, 781–791.
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical Validity of Circulating Tumour Cells in Patients with Metastatic Breast Cancer: A Pooled Analysis of Individual Patient Data. Lancet Oncol. 2014, 15, 406–414.
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells versus Imaging—Predicting Overall Survival in Metastatic Breast Cancer. Clin. Cancer Res. 2006, 12, 6403–6409.
- Pachmann, K.; Camara, O.; Kavallaris, A.; Krauspe, S.; Malarski, N.; Gajda, M.; Kroll, T.; Jörke, C.; Hammer, U.; Altendorf-Hofmann, A.; et al. Monitoring the Response of Circulating Epithelial Tumor Cells to Adjuvant Chemotherapy in Breast Cancer Allows Detection of Patients at Risk of Early Relapse. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 1208–1215.
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating Tumour Cells in Non-Metastatic Breast Cancer: A Prospective Study. Lancet Oncol. 2012, 13, 688–695.
- Smerage, J.B.; Barlow, W.E.; Hortobagyi, G.N.; Winer, E.P.; Leyland-Jones, B.; Srkalovic, G.; Tejwani, S.; Schott, A.F.; O’Rourke, M.A.; Lew, D.L.; et al. Circulating Tumor Cells and Response to Chemotherapy in Metastatic Breast Cancer: SWOG S0500. J. Clin. Oncol. 2014, 32, 3483–3489.
- Punnoose, E.A.; Atwal, S.; Liu, W.; Raja, R.; Fine, B.M.; Hughes, B.G.M.; Hicks, R.J.; Hampton, G.M.; Amler, L.C.; Pirzkall, A.; et al. Evaluation of Circulating Tumor Cells and Circulating Tumor DNA in Non–Small Cell Lung Cancer: Association with Clinical Endpoints in a Phase II Clinical Trial of Pertuzumab and Erlotinib. Clin. Cancer Res. 2012, 18, 2391–2401.
- Hou, J.-M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.C.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients With Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 525–532.
- Cohen, S.; Punt, C.; Iannotti, N.; Saidman, B.; Sabbath, K.; Gabrail, N.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Cohen SJ, Punt CJA, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LWMM, Meropol NJRelationship of Circulating Tumor Cells to Tumor Response, Progression-Free Survival, and Overall Survival in Patients with Metastatic Colorectal Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 3213–3221.
- Bork, U.; Rahbari, N.N.; Schölch, S.; Reissfelder, C.; Kahlert, C.; Büchler, M.W.; Weitz, J.; Koch, M. Circulating Tumour Cells and Outcome in Non-Metastatic Colorectal Cancer: A Prospective Study. Br. J. Cancer 2015, 112, 1306–1313.
- Kelley, R.K.; Magbanua, M.J.M.; Butler, T.M.; Collisson, E.A.; Hwang, J.; Sidiropoulos, N.; Evason, K.; McWhirter, R.M.; Hameed, B.; Wayne, E.M.; et al. Circulating Tumor Cells in Hepatocellular Carcinoma: A Pilot Study of Detection, Enumeration, and next-Generation Sequencing in Cases and Controls. BMC Cancer 2015, 15, 206.
- Okegawa, T.; Itaya, N.; Hara, H.; Tambo, M.; Nutahara, K. Circulating Tumor Cells as a Biomarker Predictive of Sensitivity to Docetaxel Chemotherapy in Patients with Castration-Resistant Prostate Cancer. Anticancer Res. 2014, 34, 6705–6710.
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.W.M.; Pienta, K.J.; Raghavan, D. Circulating Tumor Cells Predict Survival Benefit from Treatment in Metastatic Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2008, 14, 6302–6309.
- Ilie, M.; Long, E.; Butori, C.; Hofman, V.; Coelle, C.; Mauro, V.; Zahaf, K.; Marquette, C.H.; Mouroux, J.; Paterlini-Bréchot, P.; et al. ALK-Gene Rearrangement: A Comparative Analysis on Circulating Tumour Cells and Tumour Tissue from Patients with Lung Adenocarcinoma. Ann. Oncol. 2012, 23, 2907–2913.
- Xu, W.; Cao, L.; Chen, L.; Li, J.; Zhang, X.-F.; Qian, H.-H.; Kang, X.-Y.; Zhang, Y.; Liao, J.; Shi, L.-H.; et al. Isolation of Circulating Tumor Cells in Patients with Hepatocellular Carcinoma Using a Novel Cell Separation Strategy. Clin. Cancer Res. 2011, 17, 3783–3793.
- Panteleakou, Z.; Lembessis, P.; Sourla, A.; Pissimissis, N.; Polyzos, A.; Deliveliotis, C.; Koutsilieris, M. Detection of Circulating Tumor Cells in Prostate Cancer Patients: Methodological Pitfalls and Clinical Relevance. Mol. Med. 2009, 15, 101–114.
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Nakazawa, M.; Nadal, R.; Paller, C.J.; Denmeade, S.R.; Carducci, M.A.; et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015, 1, 582–591.
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441–1449.
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; McLaughlin, B.; Jendrisak, A.; Wang, Y.; Lee, J.; Greene, S.; Krupa, R.; Lu, D.; et al. Phenotypic Heterogeneity of Circulating Tumor Cells Informs Clinical Decisions between AR Signaling Inhibitors and Taxanes in Metastatic Prostate Cancer. Cancer Res. 2017, 77, 5687–5698.
- Proebstle, T.M.; Jiang, W.; Högel, J.; Keilholz, U.; Weber, L.; Voit, C. Correlation of Positive RT-PCR for Tyrosinase in Peripheral Blood of Malignant Melanoma Patients with Clinical Stage, Survival and Other Risk Factors. Br. J. Cancer 2000, 82, 118–123.
- Hoshimoto, S.; Faries, M.B.; Morton, D.L.; Shingai, T.; Kuo, C.; Wang, H.; Elashoff, R.; Mozzillo, N.; Kelley, M.C.; Thompson, J.F.; et al. Assessment of Prognostic Circulating Tumor Cells in a Phase III Trial of Adjuvant Immunotherapy After Complete Resection of Stage IV Melanoma. Ann. Surg. 2012, 255, 357.
- Alix-Panabières, C.; Pantel, K. Challenges in Circulating Tumour Cell Research. Nat. Rev. Cancer 2014, 14, 623–631.
- Swennenhuis, J.F.; van Dalum, G.; Zeune, L.L.; Terstappen, L.W.M.M. Improving the CellSearch® System. Expert Rev. Mol. Diagn 2016, 16, 1291–1305.
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating Tumor Cells: A Review of Non-EpCAM-Based Approaches for Cell Enrichment and Isolation. Clin. Chem. 2016, 62, 571–581.
- Wang, L.; Balasubramanian, P.; Chen, A.P.; Kummar, S.; Evrard, Y.A.; Kinders, R.J. Promise and Limits of the CellSearch Platform for Evaluating Pharmacodynamics in Circulating Tumor Cells. Semin. Oncol. 2016, 43, 464–475.
- De Luca, F.; Rotunno, G.; Salvianti, F.; Galardi, F.; Pestrin, M.; Gabellini, S.; Simi, L.; Mancini, I.; Vannucchi, A.M.; Pazzagli, M.; et al. Mutational Analysis of Single Circulating Tumor Cells by next Generation Sequencing in Metastatic Breast Cancer. Oncotarget 2016, 7, 26107–26119.
- Pethig, R. Review Article-Dielectrophoresis: Status of the Theory, Technology, and Applications. Biomicrofluidics 2010, 4, 022811.
- Alshareef, M.; Metrakos, N.; Juarez Perez, E.; Azer, F.; Yang, F.; Yang, X.; Wang, G. Separation of Tumor Cells with Dielectrophoresis-Based Microfluidic Chip. Biomicrofluidics 2013, 7, 011803.
- Bhagwat, N.; Dulmage, K.; Pletcher, C.H.; Wang, L.; DeMuth, W.; Sen, M.; Balli, D.; Yee, S.S.; Sa, S.; Tong, F.; et al. An Integrated Flow Cytometry-Based Platform for Isolation and Molecular Characterization of Circulating Tumor Single Cells and Clusters. Sci. Rep. 2018, 8, 5035.
- Magbanua, M.J.M.; Park, J.W. Isolation of Circulating Tumor Cells by Immunomagnetic Enrichment and Fluorescence-Activated Cell Sorting (IE/FACS) for Molecular Profiling. Methods 2013, 64, 114–118.
- Eifler, R.L.; Lind, J.; Falkenhagen, D.; Weber, V.; Fischer, M.B.; Zeillinger, R. Enrichment of Circulating Tumor Cells from a Large Blood Volume Using Leukapheresis and Elutriation: Proof of Concept. Cytom. B Clin. Cytom. 2011, 80, 100–111.
- Lee, S.-K.; Kim, G.-S.; Wu, Y.; Kim, D.-J.; Lu, Y.; Kwak, M.; Han, L.; Hyung, J.-H.; Seol, J.-K.; Sander, C.; et al. Nanowire Substrate-Based Laser Scanning Cytometry for Quantitation of Circulating Tumor Cells. Nano Lett. 2012, 12, 2697–2704.
- Mastoraki, S.; Strati, A.; Tzanikou, E.; Chimonidou, M.; Politaki, E.; Voutsina, A.; Psyrri, A.; Georgoulias, V.; Lianidou, E. ESR1 Methylation: A Liquid Biopsy-Based Epigenetic Assay for the Follow-up of Patients with Metastatic Breast Cancer Receiving Endocrine Treatment. Clin. Cancer Res. 2018, 24, 1500–1510.
- Bustin, S.A.; Siddiqi, S.; Ahmed, S.; Hands, R.; Dorudi, S. Quantification of Cytokeratin 20, Carcinoembryonic Antigen and Guanylyl Cyclase C MRNA Levels in Lymph Nodes May Not Predict Treatment Failure in Colorectal Cancer Patients. Int. J. Cancer 2004, 108, 412–417.
- ZHANG, B.; XU, C.-W.; SHAO, Y.; WANG, H.-T.; WU, Y.-F.; SONG, Y.-Y.; LI, X.-B.; ZHANG, Z.; WANG, W.-J.; LI, L.-Q.; et al. Comparison of Droplet Digital PCR and Conventional Quantitative PCR for Measuring EGFR Gene Mutation. Exp. Med. 2015, 9, 1383–1388.
- Siravegna, G.; Bardelli, A. Minimal Residual Disease in Breast Cancer: In Blood Veritas. Clin. Cancer Res. 2014, 20, 2505–2507.
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610.
- Soler, A.; Cayrefourcq, L.; Mazel, M.; Alix-Panabières, C. EpCAM-Independent Enrichment and Detection of Viable Circulating Tumor Cells Using the EPISPOT Assay. Methods Mol. Biol. 2017, 1634, 263–276.
- Eyer, K.; Doineau, R.C.L.; Castrillon, C.E.; Briseño-Roa, L.; Menrath, V.; Mottet, G.; England, P.; Godina, A.; Brient-Litzler, E.; Nizak, C.; et al. Single-Cell Deep Phenotyping of IgG-Secreting Cells for High-Resolution Immune Monitoring. Nat. Biotechnol. 2017, 35, 977–982.
- Millner, L.M.; Linder, M.W.; Valdes, R. Circulating Tumor Cells: A Review of Present Methods and the Need to Identify Heterogeneous Phenotypes. Ann. Clin. Lab. Sci 2013, 43, 295–304.
- Ramirez, J.-M.; Fehm, T.; Orsini, M.; Cayrefourcq, L.; Maudelonde, T.; Pantel, K.; Alix-Panabières, C. Prognostic Relevance of Viable Circulating Tumor Cells Detected by EPISPOT in Metastatic Breast Cancer Patients. Clin. Chem. 2014, 60, 214–221.
- Monteiro-Riviere, N.A.; Inman, A.O.; Zhang, L.W. Limitations and Relative Utility of Screening Assays to Assess Engineered Nanoparticle Toxicity in a Human Cell Line. Toxicol. Appl. Pharm. 2009, 234, 222–235.
- Catelain, C.; Pailler, E.; Oulhen, M.; Faugeroux, V.; Pommier, A.-L.; Farace, F. Detection of Gene Rearrangements in Circulating Tumor Cells: Examples of ALK-, ROS1-, RET-Rearrangements in Non-Small-Cell Lung Cancer and ERG-Rearrangements in Prostate Cancer. In Isolation and Molecular Characterization of Circulating Tumor Cells; Advances in Experimental Medicine and, Biology; Magbanua, M.J.M., Park, J.W., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 169–179. ISBN 978-3-319-55947-6.
- Carter, L.; Rothwell, D.G.; Mesquita, B.; Smowton, C.; Leong, H.S.; Fernandez-Gutierrez, F.; Li, Y.; Burt, D.J.; Antonello, J.; Morrow, C.J.; et al. Molecular Analysis of Circulating Tumor Cells Identifies Distinct Copy-Number Profiles in Patients with Chemosensitive and Chemorefractory Small-Cell Lung Cancer. Nat. Med. 2017, 23, 114–119.
- Lohr, J.G.; Adalsteinsson, V.A.; Cibulskis, K.; Choudhury, A.D.; Rosenberg, M.; Cruz-Gordillo, P.; Francis, J.M.; Zhang, C.-Z.; Shalek, A.K.; Satija, R.; et al. Whole-Exome Sequencing of Circulating Tumor Cells Provides a Window into Metastatic Prostate Cancer. Nat. Biotechnol. 2014, 32, 479–484.
- Babayan, A.; Alawi, M.; Gormley, M.; Müller, V.; Wikman, H.; McMullin, R.P.; Smirnov, D.A.; Li, W.; Geffken, M.; Pantel, K.; et al. Comparative Study of Whole Genome Amplification and next Generation Sequencing Performance of Single Cancer Cells. Oncotarget 2017, 8, 56066–56080.
- Harb, W.; Fan, A.; Tran, T.; Danila, D.C.; Keys, D.; Schwartz, M.; Ionescu-Zanetti, C. Mutational Analysis of Circulating Tumor Cells Using a Novel Microfluidic Collection Device and QPCR Assay. Transl. Oncol. 2013, 6, 528–538.
- Ting, D.T.; Wittner, B.S.; Ligorio, M.; Jordan, N.V.; Shah, A.M.; Miyamoto, D.T.; Aceto, N.; Bersani, F.; Brannigan, B.W.; Xega, K.; et al. Single-Cell RNA Sequencing Identifies Extracellular Matrix Gene Expression by Pancreatic Circulating Tumor Cells. Cell Rep. 2014, 8, 1905–1918.
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515.
- Jordan, N.V.; Bardia, A.; Wittner, B.S.; Benes, C.; Ligorio, M.; Zheng, Y.; Yu, M.; Sundaresan, T.K.; Licausi, J.A.; Desai, R.; et al. HER2 Expression Identifies Dynamic Functional States within Circulating Breast Cancer Cells. Nature 2016, 537, 102–106.
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238.
- Plagnol, V.; Woodhouse, S.; Howarth, K.; Lensing, S.; Smith, M.; Epstein, M.; Madi, M.; Smalley, S.; Leroy, C.; Hinton, J.; et al. Analytical Validation of a next Generation Sequencing Liquid Biopsy Assay for High Sensitivity Broad Molecular Profiling. PLoS ONE 2018, 13, e0193802.
- Bobek, V.; Gurlich, R.; Eliasova, P.; Kolostova, K. Circulating Tumor Cells in Pancreatic Cancer Patients: Enrichment and Cultivation. World J. Gastroenterol. 2014, 20, 17163–17170.
- Chinen, L.T.D.; Mello, C.A.L.; Abdallah, E.A.; Ocea, L.M.; Buim, M.E.; Breve, N.M.; Gasparini, J.L.; Fanelli, M.F.; Paterlini-Bréchot, P. Isolation, Detection, and Immunomorphological Characterization of Circulating Tumor Cells (CTCs) from Patients with Different Types of Sarcoma Using Isolation by Size of Tumor Cells: A Window on Sarcoma-Cell Invasion. Onco Targets Ther. 2014, 7, 1609–1617.
- Nature Communications. Profiling Protein Expression in Circulating Tumour Cells Using Microfluidic Western Blotting. Available online: https://www.nature.com/articles/ncomms14622 (accessed on 21 December 2022).
- Baselga, J. Targeting the Phosphoinositide-3 (PI3) Kinase Pathway in Breast Cancer. Oncologist 2011, 16 (Suppl. 1), 12–19.
- Alix-Panabières, C.; Cayrefourcq, L.; Mazard, T.; Maudelonde, T.; Assenat, E.; Assou, S. Molecular Portrait of Metastasis-Competent Circulating Tumor Cells in Colon Cancer Reveals the Crucial Role of Genes Regulating Energy Metabolism and DNA Repair. Clin. Chem. 2017, 63, 700–713.
- Soler, A.; Cayrefourcq, L.; Mazard, T.; Babayan, A.; Lamy, P.-J.; Assou, S.; Assenat, E.; Pantel, K.; Alix-Panabières, C. Autologous Cell Lines from Circulating Colon Cancer Cells Captured from Sequential Liquid Biopsies as Model to Study Therapy-Driven Tumor Changes. Sci. Rep. 2018, 8, 15931.
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a Population of Blood Circulating Tumor Cells from Breast Cancer Patients That Initiates Metastasis in a Xenograft Assay. Nat. Biotechnol. 2013, 31, 539–544.
- Hodgkinson, C.L.; Morrow, C.J.; Li, Y.; Metcalf, R.L.; Rothwell, D.G.; Trapani, F.; Polanski, R.; Burt, D.J.; Simpson, K.L.; Morris, K.; et al. Tumorigenicity and Genetic Profiling of Circulating Tumor Cells in Small-Cell Lung Cancer. Nat. Med. 2014, 20, 897–903.
More
