Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Peter Tang and Version 1 by Hui Cao.
RNA-binding proteins (RBPs) are key regulators of transcription and translation, with highly dynamic spatio-temporal regulation. They are usually involved in the regulation of RNA splicing, polyadenylation, and mRNA stability and mediate processes such as mRNA localization and translation, thereby affecting the RNA life cycle and causing the production of abnormal protein phenotypes that lead to tumorigenesis and development. Accumulating evidence supports that RBPs play critical roles in vital life processes, such as bladder cancer initiation, progression, metastasis, and drug resistance.
- RNA binding proteins
- bladder cancer
- LIN28B
- human antigen R
- heterogeneous nuclear RNPs
1. Introduction
Tumor formation in humans is an extremely complex and multi-stage process that typically occurs over years or decades. The histiocytes of normal individuals gradually develop into tumors with malignant phenotypes through evolution, through a process called tumor progression. Tumors can occur in various tissues of the human body, and the incidence increases with age. Only in very rare cases do cancerous cells progress to clinically visible tumor tissue with occupying lesions. Tumor progression is closely related to epigenetics, RNA post-transcriptional modification, protein post-translational modification, and other life processes. Tumors are not only regulated by these life processes but are also affected by normal biochemical reactions, reshaping cellular life activities and ultimately giving cells the ability to proliferate indefinitely.
Bladder cancer is the commonest malignant tumor in the urinary system. According to cancer statistics released in 2023, the estimated incidence of bladder cancer in the United States is 82,290 cases, and the mortality rate is 16,170 cases [1]. The incidence is higher than that in 2022, while the mortality is slightly lower than that in 2022 [2]. According to the report published by China, the incidence of bladder cancer in 2020 was 91,893 cases, with mortality in 42,973 cases [3]. Bladder cancer is a broad concept that encompasses everything from low-risk non-muscle-invasive bladder cancer to high-risk primary invasive bladder cancer. Low- and intermediate-risk non-muscle invasive bladder cancer (NMIBC) patients face high recurrence rates, with 5-year event-free survival rates reaching 43% and 33% [4]. Metastasis of bladder cancer is a catastrophe that 50–70% of muscle invasive bladder cancer (MIBC) patients have to face, and given the extremely high metastasis rate, the 5-year overall survival rate for advanced MIBC is 4.8% [5].
RBPs play crucial roles in the regulation of cellular life processes, especially RNA splicing, modification, transport, localization, stabilization, degradation, and translation. Certain RBPs are expressed in a variety of cells to maintain essential cellular functions. Altered structure or disturbed expression of RBPs may cause different diseases, and this concept is reflected in tumorigenesis [6]. Given that RBPs can regulate post-transcriptional RNA, they can rapidly and efficiently alter gene expression in response to changes in the microenvironment. A single RBP can bind multiple targets, and different combinations of several RNP interactions contribute to enhanced cellular recognition and responses to stress [7]. In addition, RBP can promote mRNA translation by recruiting specific translation signaling molecules [8]. By contrast, RBPs involved in the RNA-induced silencing complex can inhibit target mRNA translation while inducing its degradation [9,10][9][10].
2. RNA Binding Motif 3 (RBM3)
RBM3 is a glycine-rich cold shock protein whose expression can be stimulated by hypothermia, ischemia, or hypoxia [22,23,24][11][12][13]. RBM3 has two highly conserved RRMs, namely RNP1 and RNP2, at the N terminus and an arginine-glycine-rich domain (RGG) at the C terminus [16][14]. The RGG structural domain mainly regulates the process of RNA cleavage and polyadenylate cyclization [25][15]. The RGG structural domain, especially the part with arginine residues, is essential for mRNA export, because the absence of a single arginine residue in the RGG structural domain can interrupt the shuttling process of RBM3 between the nucleus and cytoplasm [25,26][15][16]. RBM3 performs four main functions in tumors. RBM3 can bind and affect the translation of mRNA. It influences mRNA stability and the translation of COX-2, interleukin (IL)-8, and vascular endothelial growth factor (VEGF) [27][17]. In general, RBM3 facilitates the translation of various mRNAs into proteins [26,27,28][16][17][18]. This promotion includes the following mechanisms: (1) binding to the 60S ribosomal subunit in an RNA-independent manner [28][18]; (2) increased formation of active polyribosomes [27][17]; (3) dephosphorylation of eukaryotic initiation factor (eIF2α); (4) promotion of eIF4E phosphorylation [28][18]. Under low-temperature conditions, RBM3 can alter miRNA levels and thus promote protein translation. RBM3 binds to a precursor miRNA and facilitates its processing by the Dicer complex to form a mature double-stranded miRNA [29][19]. The regulation of miRNAs by RBM3 is two-sided: it can positively regulate most miRNAs, but reducing the level of RBM3 can promote the expression of a small number of temperature-sensitive miRNAs, thereby preventing pathological hyperthermia [30][20]. These results suggest that RBM3 is essential for the mitotic process of cells. RBM3 can play a regulatory role in the cell cycle of G2/M transition. In colorectal cancer cells, RBM3 induces stem cell proliferation through a mechanism that increases β-catenin signaling by inhibiting glycogen synthase kinase-3 beta kinase activity [31][21]. By contrast, knockdown of RBM3 expression in the human HCT116 colon cancer cell line caused increases in caspase-dependent apoptosis, nuclear cyclin B1 expression, and Cdc25c, Chk1, and Chk2 phosphorylation levels, which are a series of alterations suggesting that downregulation of RBM3 will prevent cell mitosis [27][17]. In vivo, embryonic fibroblasts from RBM3-deficient mice showed a significant increase in the number of cells in the G2 phase [32][22]. These conclusions can also explain the higher sensitivity of tumors with a high RBM3 expression to chemotherapy than those with low or negative expression [33][23]. When cells receive various external stimuli, unfolded proteins accumulate in the lumen of the endoplasmic reticulum and activate the unfolded protein response (UPR) to rescue cells. Sustained and/or intense endoplasmic reticulum stress (ERS) induces apoptosis [34][24]. The protein kinase R-like endoplasmic reticulum kinase (PERK)-eIF2α-C/EBP-homologous protein (CHOP) signaling pathway plays an important role in UPR-induced apoptosis [35][25]. Under sustained ERS, RBM3 can inhibit the phosphorylation of PERK and eIF2α, causing a decrease in the expression of CHOP and inhibiting UPR to avoid apoptosis [36][26]. This condition may be the reason why the UPR does not induce apoptosis despite its low-temperature activation; low-temperature-induced RBM3 may play an important role in this process [37][27]. Hypothermia can also alleviate ischemia-induced apoptosis by inhibiting the UPR [38][28]. In bladder cancer, the role of RBM3 is similar to that in other tumors. A clinical retrospective study including 259 bladder cancer patients showed that the low expression of RBM3 was an independent factor for poor prognosis of bladder cancer [39][29]; this finding is closely related to the progression of bladder cancer and decreased overall survival of patients [40][30]. A similar study revealed that patients with a high expression of RBM3 were associated not only with a low tumor grade but also with a low risk of lymphovascular invasion (lymph node invasion) [41][31]. The effect of RBM3 on bladder cancer may depend on several factors: (1) the expression level of RBM3 is closely related to tumor stage; (2) RBM3 silencing can increase the number of G2/M stage cells and eventually lead to apoptosis [33][23]; (3) RBM3 directly binds to a variety of mRNAs, thus regulating the activity of multiple kinases in tumors [42,43][32][33].3. LIN28
LIN28 is a highly conserved RNA-binding protein in eukaryotes [44][34]. In a variety of mammals, including humans, LIN28 is divided into LIN28A and its homologous molecule, LIN28B [45][35]. Human LIN28A is encoded by the Lin28a gene, which is located on chromosome 1p36.11 and is mainly expressed in embryonic stem cells and embryonic carcinoma cells [46,47][36][37]. LIN28B is encoded by the Lin28b gene on chromosome 6q21 and is mainly expressed in the testis, placenta, and other tissues [45,48][35][38]. LIN28A and LIN28B have highly similar protein structures: both have two functional domains, namely a cold shock protein domain (CSD) and a retroviral zinc finger (cys-cys-his-cys, CCHC) domain [46][36]. After mutation of either domain, the other domain still has the function of binding RNA, suggesting that CSD and CCHC can participate in the RNA binding of LIN28 [49][39]. LIN28 protein is mainly localized in the cytoplasm [47][37]. However, it can also be present in RNPs, polyribosomes (polysome), P vesicles, and stress granules [50][40]. Meanwhile, LIN28B is mainly located in the nucleus, and it may exert its biological function through the cytoplasmic microprocessor [51][41]. However, the expressions of LIN28A and LIN28B are mutually exclusive, and tumor cells expressing LIN28A do not express LIN28B, and vice versa [51][41]. The miRNA let-7 family contains 12 miRNA members, which act as tumor suppressors and inhibit the expression of a variety of important oncogenes (including Ras, Myc, and so on) by binding to their 3′ untranslated regions [52,53,54][42][43][44]. This function of let-7 is regulated by the RNA-binding protein LIN28 [55][45]. Overexpression of LIN28A or LIN28B is associated with a variety of tumors, leading to increased tumor aggressiveness and poorer prognosis [56][46]. LIN28B has also attracted considerable attention as one of the downstream genes of nuclear factor (NF)-κB [57][47]. LIN28A and LIN28B can inhibit the expression of oncogenes, such as Ras and Myc, by inhibiting let-7-miRNA [58,59][48][49]. MiRNA let-7 acts as the main effector molecule of LIN28A and LIN28B, with which they form multiple feedback loops: (1) LIN28A/B can inhibit the maturation of let-7 through various mechanisms, whereas let-7 can inhibit the translation of LIN28A/B at the post-transcriptional level, reducing the protein expression level [60][50]; (2) after LIN28A/B inhibits the maturation of let-7, the inhibition of c-Myc by let-7 is relieved, and c-Myc can promote the transcription of LIN28A/B, forming a positive feedback loop [61,62][51][52]; (3) NF-κB can induce the expression of LIN28B, and LIN28B inhibits the maturation of let-7, thus releasing the inhibitory effect of let-7 on the expression of IL-6, which can activate the expression of NF-κB, forming a positive feedback loop. Thus, linking inflammation and tumor further reveals the role played by inflammatory factors in the malignant transformation of cells (Figure 1) [63][53].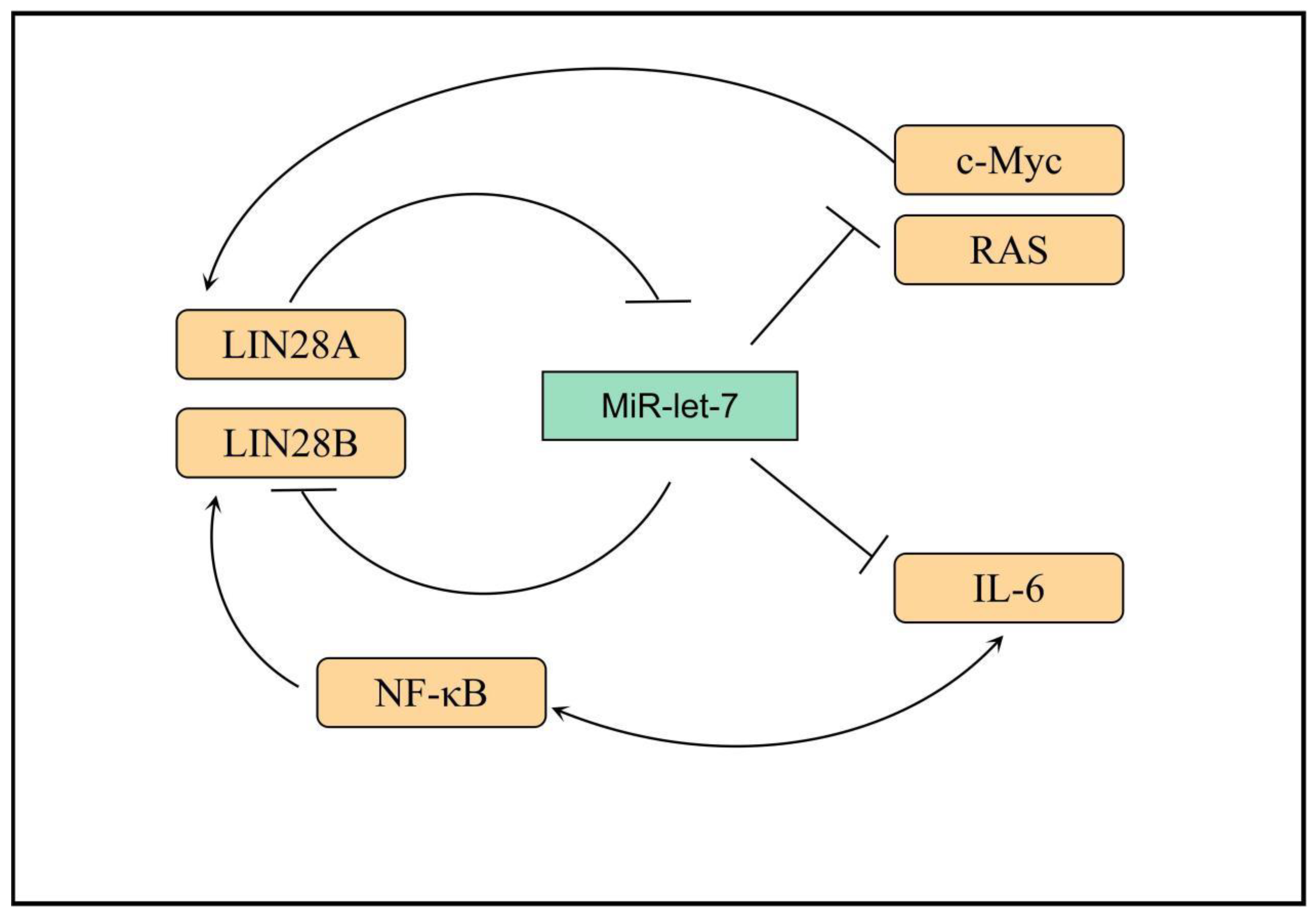
Figure 1. Transcriptional networks that regulate LIN28B expression. LIN28B expression is lost. In adult mammals, only a small subset of somatic cells exist where LIN28B expression occurs. Several transcription factors, such as MYC and NF-κB, promote LIN28B transcription, while REST and ESE3/EHF are transcriptional repressors. IL-6: interleukin-6, RAS: Resistance to audiogenic seizures, NF-κB: nuclear factor kappa-B, MYC: MYC protooncogene.
4. HuR
HuR is an embryonic lethal abnormal vision gene that includes four family members: HuB, HuC, HuD, and HuR. The first three are expressed mainly in neural tissues and germ cells and are associated with neurodevelopment, whereas HuR is commonly expressed in all human cells [71][54]. The human HuR gene (hHuR) is located on chromosome 19p13.2, which is closely related to chromosomal translocations and tumor carcinogenesis in human tumors [72][55]. HuR contains three RRMs and a hinge region in which RRM1 and RRM2 bind to adenine- and uracil-rich elements (AU-rich elements, AREs) in the target mRNA. By contrast, RRM3 can bind to the polyadenylate tail of the target mRNA. In normal conditions, AREs can accelerate the poly-A tail of mRNA to undergo deadenylation to destabilize mRNA [73][56]. Therefore, when HuR protein binds to these AREs, it can inhibit its own deadenylation and help mRNA to be protected from nuclease degradation during mRNA transport from nucleus to cytoplasm, thereby increasing mRNA stability and promoting mRNA translation; thus, HuR plays a role in post-transcriptional regulation [73,74][56][57]. The hinge region between RRM2 and RRM3 contains a 52-amino-acid HuR nucleoplasmic shuttling sequence, which is the main motif for post-translational modification of HuR and a key region for nucleoplasmic transport (Figure 2) [75,76][58][59].
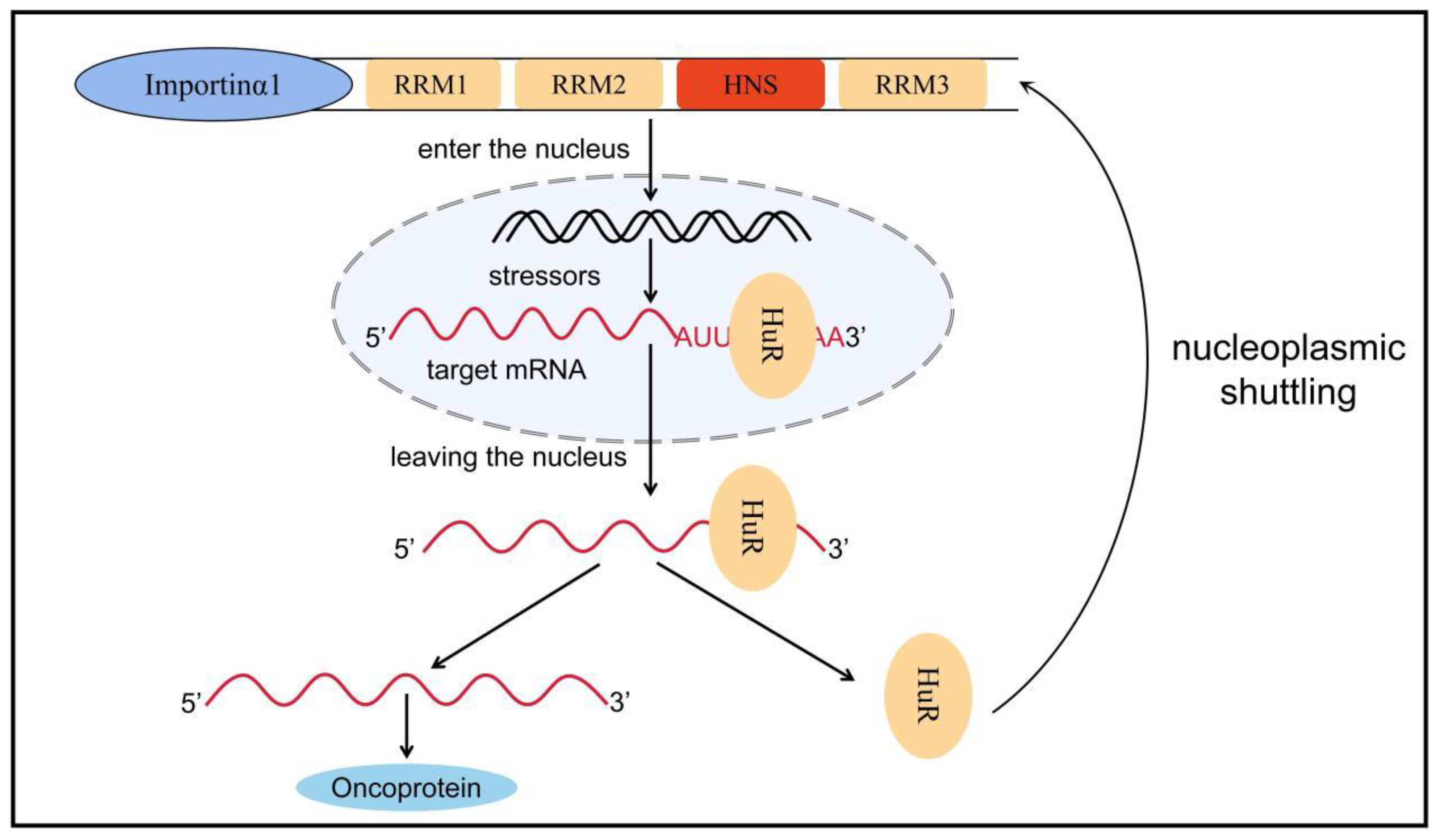

Figure 2. Nucleoplasmic transport of HuR. HuR is mainly distributed in the nucleus, but when stimulated by factors such as microenvironmental changes, HuR can bind to target mRNA to form a HuR-mRNA complex, which protects the target mRNA from degradation by nucleic acid exonucleases and transfers it from the nucleus to the cytoplasm via HNS. Subsequently, HuR dissociates from the target mRNA and rapidly returns to the nucleus with the assistance of transporter proteins such as importin α1. Nucleoplasmic translocation of HuR increases the stability of the target mRNA, promotes mRNA translation, and causes a variety of inflammatory phenotypes, ultimately promoting tumor formation and progression. RRM: RNA recognition motif, HuR: human antigen R, HNS: HuR nucleoplasmic shuttling sequence.
5. Heterogeneous Nuclear RNPs (hnRNPs)
HnRNPs act as RBPs by binding to pre-mRNA to form the hnRNP–RNA complex. Subsequently, they become involved in the processes of mRNA splicing, translation, transport, and biodegradation [105][60]. HnRNPs can be further subdivided into several subgroups, named in order from A to U, with relative molecular weights ranging from 34,000 kD to 120,000 kD [106,107][61][62]. HnRNPs have four unique RBDs: RRMs, quasi-RRM, the arginine glycine cluster (RGG box), and the nuclear protein KH structural domain [108,109,110][63][64][65]. Two highly similar RRMs form a βαββαβ structure in eukaryotic cells, and they contain two highly conserved shared RNP sequences [111][66]. Although these RRMs are highly similar, a significant difference exists in their affinity for RNA binding. In most cases, the RRMs preferentially bind RNA and can recognize longer motifs, whereas quasi-RRMs bind weakly and mainly assist in the binding of proteins to RNA. However, the disruption of RRM interactions or loss of either binding capacity can affect the function of hnRNPs [112][67]. The RGG region is the main auxiliary region of hnRNPs and mainly mediates the interaction of homologous or heterologous proteins with hnRNPs [110][65]. The KH domain forms the structure of βααββα, whose function is related to the splicing of target mRNAs [113][68]. In addition, several hnRNAs have a nucleoplasmic shuttle function, and they can form complexes with pre-mRNAs to assist mRNAs in the nucleoplasmic transport process (Figure 3) [114][69]. The M9 sequence is a special class of nucleoplasmic shuttle sequence that is distinct from the traditional NLS and is responsible for the bidirectional regulation of the transport of hnRNPs with shuttle function from the nucleus to the cytoplasm [115][70]. The nucleocytoplasmic shuttling function of hnRNPs relies on the complete M9 sequence. Single-amino-acid site mutations can disrupt normal protein input and output processes [116][71]. A part of the hnRNP also has several auxiliary sequences, such as Gly- and Pro-rich domains, which mediate protein–protein interactions, subcellular localization, and other functions [106][61]. Table 1 shows the structural and functional characteristics of the main members of the hnRNP family.
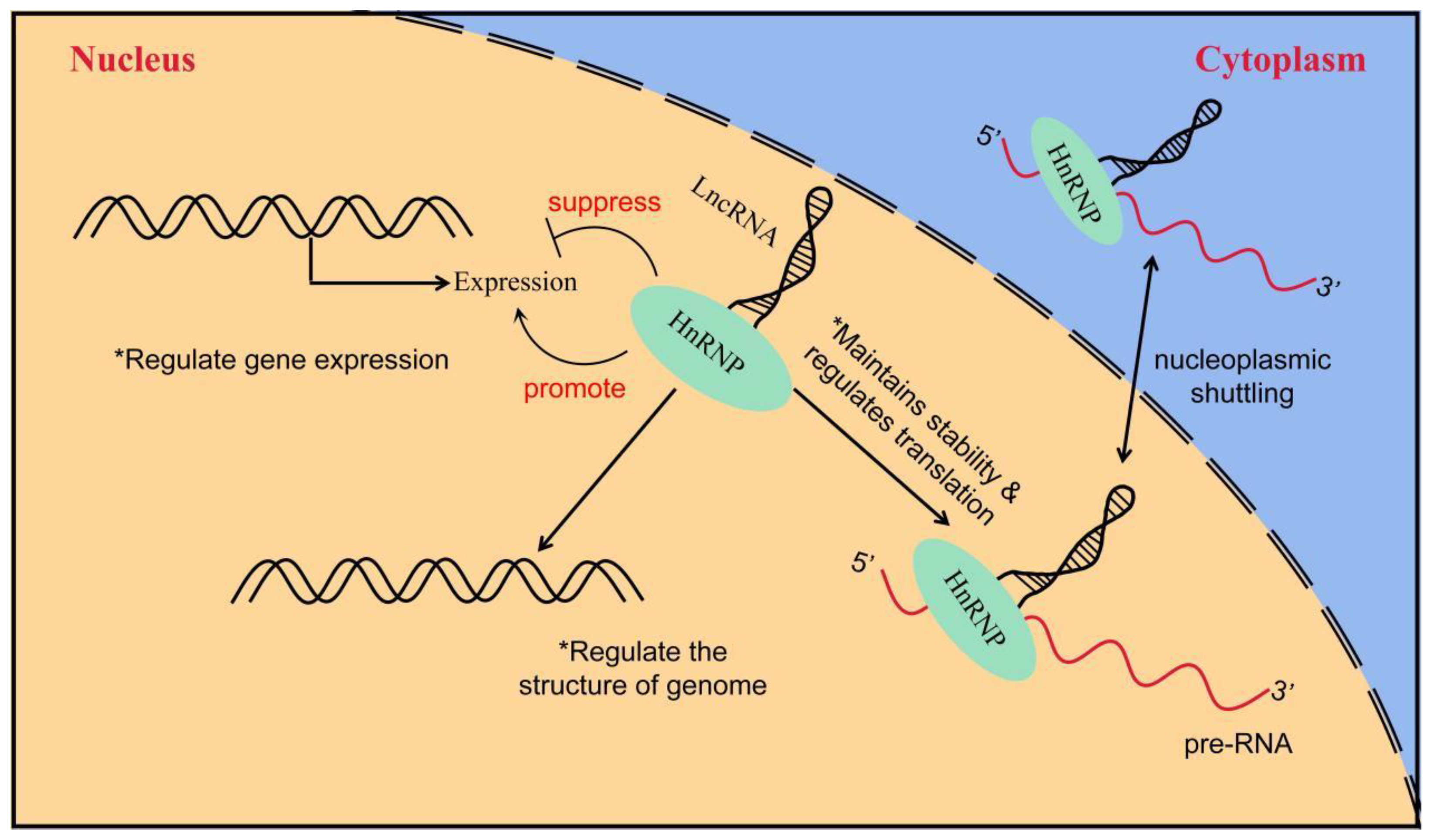

Figure 3. The way in which LncRNA interacts with hnRNP to regulate gene expression. LncRNA interacts with hnRNP to induce or inhibit gene expression; LncRNA regulates genome structure through interaction with hnRNP and indirectly affects gene expression; the interaction between lncRNA and hnRNP controls the stability and translation of mRNA.
6. Others
Although four types of RBPs have been listed for their roles and functions in bladder cancer, more RBPs have been identified and reported in bladder cancer, including insulin-like growth factor messenger RNA binding protein 3 (IGF2BP3), nucleolin (NCL), and quaking (QKI). Moreover, it is reported that Fragile X-related gene 1 (FXR1) was identified as a novel cancer driver gene in urothelial carcinoma of the bladder (UCB) [132][72]; circ-SLC38A1 promotes BC cells invasion in vitro and lung metastasis in vivo in mice [133][73].
6.1. IGF2BP3
IGF2BP3 is a member of the IGF2BP family, and its members all contain 2 RRM and 4 KH domains in their structure; these 4 KH domains are all binding GGC sequences [134][74]. Under normal or stress conditions, members of the IGF2BP family can act on target mRNAs in an N6-methyladenosine-dependent manner, promote their stability, and increase their intracellular content, thereby affecting gene expression [135,136][75][76]. However, currently, only IGF2BP3 has been reported in bladder cancer. The chromosomal localization of IGF2BP3 is 7p15.3.IGF2BP3, which is a typical multi-domain RNA-binding protein with specificity and diversity in recognizing targets and provides a good paradigm for the multivalent interactions of multi-domain RNA-binding proteins [137][77].
Compared with IGF2BP3, which has been widely reported in a variety of tumors such as breast, liver, and gastrointestinal tract tumors, pancreatic cancer, and lung cancer [138][78], knowledge on its role in bladder cancer is limited. Regardless, IGF2BP3 is highly expressed in bladder cancer patients and independently associated with bladder cancer recurrence, cancer-specific mortality, and all-cause mortality [139][79]. Knocking down the expression of IGF2BP3 in bladder cancer can increase apoptosis and induce cell cycle arrest, implying that it originally promoted cell proliferation via inhibiting apoptosis and regulating cell cycle [140][80].
6.2. NCL
NCL is one of the abundant proteins in the nucleolus, and it is widely distributed in the nucleolus, nucleoplasm, cytoplasm, and cell membrane of eukaryotic cells, participating in a variety of biological processes [141][81]. NCL is mainly distributed in the nucleus, is responsible for the transcription of rDNA, and has various roles in the biogenesis of ribosomes, including RNA polymerase I transcription, pre-rRNA processing, and ribosome assembly [142][82]. The human NCL gene is a haploid genome located on 2q12-qter, consisting of 14 exons and 13 introns [143][83]. The N-terminal domain of the NCL is majorly involved in DNA regulation and protein-to-protein reactions, and this region also contains a variety of highly phosphorylated sites that can be involved in cell cycle regulation [143][83]. The central domain holds four RRMs that bind to and regulate the transcription of specific mRNAs [144][84]. The C-terminal sequence and arrangement of NCL is not conserved, and its length is variable, usually interspersed with a large number of glycine, arginine, and phenylalanine residues; its main function is to help NCL interact during larger or more complex RNA localization [145][85]. NCL is highly expressed in a variety of tumors [146][86]; besides promoting tumor proliferation [147[87][88][89],148,149], it is an important anti-apoptotic protein that maintains tumor cell survival [143,150,151,152,153][83][90][91][92][93]. NCL has also been reported to be involved in various processes, such as angiogenesis [154[94][95][96],155,156], infiltration, and metastasis [157,158,159,160][97][98][99][100] of tumors.
In bladder cancer, increased expression of NCL boosts its aggressiveness and promotes its pulmonary metastasis [161][101], whereas blocking NCL expression in bladder cancer can inhibit bladder cancer proliferation and invasion [162,163,164][102][103][104]. However, the target mRNAs of NCL in bladder cancer, such as Rho factor [161,162][101][102] and/or MMP-2 [163][103], are also diverse. In addition, NCL has the potential to promote tumor proliferation by promoting the function of epidermal growth factor receptor [164,165][104][105]. The role played by NCL in bladder cancer is far less clearly explained than other tumors, and this area needs more research input. Regardless, the treatment targeting NCL has great potential in bladder cancer.
6.3. QKI
QKI protein is a subfamily of the signal transduction and activation of RNA (STAR) family [166][106]. The STAR protein family plays an important role in embryogenesis, tissue, and organ development [167,168][107][108]. The human Qki gene is localized on chromosome 6 and has nine exons, which can produce at least five transcripts by different splicing methods [169][109]. Three of the main transcripts are the most important, as they are named Qki-6, Qki-6, and QkiI-7 due to their sizes (5, 6, and 7 kb, respectively). The structure of QKI proteins is highly homologous to other members of the STAR family, with similar domains: an RRM (KH domain) flanked by QUA1 and QUA2 domains [169][109]. The QKI-5 protein is mainly localized in the nucleus, where it can bind to target mRNAs and retain them in the nucleus. This condition is possible because QKI-5 contains a nuclear localization signal peptide at its C-terminus, whereas QKI-6 and QKI-7, which lack this signal peptide, are mainly localized in the cytoplasm and are involved in target mRNA transport and regulate its stability [170][110]. The reason why QKI proteins can bind to target mRNAs is that these mRNAs have a sequence that can be specifically bound by QKI, namely 5′-A(C/A)UAA-3′; hence, this sequence is also called a quaking response element (QRE) [171][111]. Bioinformatic analysis showed that QKI can interact with more than 1000 mRNAs with QRE, and most of these are important molecules in cell-directed differentiation, proliferation, metastasis, and apoptosis [171,172][111][112]. QKI has a low expression in a variety of malignancies, and overexpression of QKI inhibits the proliferative, invasive, and migratory capacities of these tumors and promotes their apoptosis [173,174,175,176,177][113][114][115][116][117]. Thus, QKI may play an important role as an oncogenic factor in the development of malignant tumors.
In bladder cancer, the low expression of QKI is associated with advanced tumor TNM staging and poor overall survival, whereas its overexpression inhibits the ability of bladder cancer cells to grow and invade [178][118]. The oncogenic mechanism of miRNA-362-5p in bladder cancer is also related to QKI, which can promote the proliferative and invasive effects of bladder cancer via targeting binding and reducing QKI [179][119]. Cancer-associated fibroblasts (CAFs) are also an important component of the tumor microenvironment, secreting microfibrillar-associated protein 5 (MFAP5), a component of elastic microfibers, which is also an oncogenic factor in a variety of tumors [180][120], promoting tumor cell proliferation, invasion [181][121], and recruiting new blood vessels [182][122]. In bladder cancer, QKI can directly target binding to MFAP5 in CAFs and downregulate its expression, thus limiting tumorigenesis and progression [183][123]. The expression level of QKI in malignant tumors can be used as an important indicator to predict the occurrence and development of cancer, and it is expected to become a new target for tumor therapy, including that of bladder cancer. However, how to apply it effectively and precisely in clinical diagnosis and treatment needs further research. In particular, the functional differences between the subtypes of QKI in tumorigenesis development are rarely reported.
References
- Siegel, R.; Miller, K.; Wagle, N.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48.
- Siegel, R.; Miller, K.; Fuchs, H.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33.
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590.
- Ritch, C.; Velasquez, M.; Kwon, D.; Becerra, M.; Soodana-Prakash, N.; Atluri, V.; Almengo, K.; Alameddine, M.; Kineish, O.; Kava, B.; et al. Use and Validation of the AUA/SUO Risk Grouping for Nonmuscle Invasive Bladder Cancer in a Contemporary Cohort. J. Urol. 2020, 203, 505–511.
- Alfred Witjes, J.; Lebret, T.; Compérat, E.; Cowan, N.; De Santis, M.; Bruins, H.; Hernández, V.; Espinós, E.; Dunn, J.; Rouanne, M.; et al. Updated 2016 EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer. Eur. Urol. 2017, 71, 462–475.
- Wang, Z.; Li, B.; Luo, Y.; Lin, Q.; Liu, S.; Zhang, X.; Zhou, H.; Yang, J.; Qu, L. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018, 22, 286–298.
- Smith, C.; Valcárcel, J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 2000, 25, 381–388.
- Michlewski, G.; Sanford, J.; Cáceres, J. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 2008, 30, 179–189.
- Fabian, M.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379.
- Kim, H.; Kuwano, Y.; Srikantan, S.; Lee, E.; Martindale, J.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748.
- Kishore, S.; Luber, S.; Zavolan, M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief. Funct. Genomics 2010, 9, 391–404.
- Phadtare, S.; Alsina, J.; Inouye, M. Cold-shock response and cold-shock proteins. Curr. Opin. Microbiol. 1999, 2, 175–180.
- Zhu, X.; Bührer, C.; Wellmann, S. Cold-inducible proteins CIRP and RBM3, a unique couple with activities far beyond the cold. Cell. Mol. Life Sci. CMLS 2016, 73, 3839–3859.
- Zhou, R.; Lu, X.; Zhang, C.; Yin, D. RNA binding motif protein 3: A potential biomarker in cancer and therapeutic target in neuroprotection. Oncotarget 2017, 8, 22235–22250.
- Godin, K.; Varani, G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007, 4, 69–75.
- Smart, F.; Aschrafi, A.; Atkins, A.; Owens, G.; Pilotte, J.; Cunningham, B.; Vanderklish, P. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J. Neurochem. 2007, 101, 1367–1379.
- Sureban, S.; Ramalingam, S.; Natarajan, G.; May, R.; Subramaniam, D.; Bishnupuri, K.; Morrison, A.; Dieckgraefe, B.; Brackett, D.; Postier, R.; et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene 2008, 27, 4544–4556.
- Dresios, J.; Aschrafi, A.; Owens, G.; Vanderklish, P.; Edelman, G.; Mauro, V. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. USA 2005, 102, 1865–1870.
- Pilotte, J.; Dupont-Versteegden, E.; Vanderklish, P. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS ONE 2011, 6, e28446.
- Wong, J.; Au, A.; Gao, D.; Pinello, N.; Kwok, C.; Thoeng, A.; Lau, K.; Gordon, J.; Schmitz, U.; Feng, Y.; et al. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. 2016, 44, 2888–2897.
- Venugopal, A.; Subramaniam, D.; Balmaceda, J.; Roy, B.; Dixon, D.; Umar, S.; Weir, S.; Anant, S. RNA binding protein RBM3 increases β-catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol. Carcinog. 2016, 55, 1503–1516.
- Matsuda, A.; Ogawa, M.; Yanai, H.; Naka, D.; Goto, A.; Ao, T.; Tanno, Y.; Takeda, K.; Watanabe, Y.; Honda, K.; et al. Generation of mice deficient in RNA-binding motif protein 3 (RBM3) and characterization of its role in innate immune responses and cell growth. Biochem. Biophys. Res. Commun. 2011, 411, 7–13.
- Ehlén, Å.; Nodin, B.; Rexhepaj, E.; Brändstedt, J.; Uhlén, M.; Alvarado-Kristensson, M.; Pontén, F.; Brennan, D.; Jirström, K. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl. Oncol. 2011, 4, 212–221.
- Nie, Z.; Chen, M.; Wen, X.; Gao, Y.; Huang, D.; Cao, H.; Peng, Y.; Guo, N.; Ni, J.; Zhang, S. Endoplasmic Reticulum Stress and Tumor Microenvironment in Bladder Cancer: The Missing Link. Front. Cell Dev. Biol. 2021, 9, 683940.
- Lin, J.; Li, H.; Yasumura, D.; Cohen, H.; Zhang, C.; Panning, B.; Shokat, K.; Lavail, M.; Walter, P. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007, 318, 944–949.
- Zhu, X.; Zelmer, A.; Kapfhammer, J.; Wellmann, S. Cold-inducible RBM3 inhibits PERK phosphorylation through cooperation with NF90 to protect cells from endoplasmic reticulum stress. FASEB J. 2016, 30, 624–634.
- Rzechorzek, N.; Connick, P.; Patani, R.; Selvaraj, B.; Chandran, S. Hypothermic Preconditioning of Human Cortical Neurons Requires Proteostatic Priming. EBioMedicine 2015, 2, 528–535.
- Poone, G.; Hasseldam, H.; Munkholm, N.; Rasmussen, R.; Grønberg, N.; Johansen, F. The Hypothermic Influence on CHOP and Ero1-α in an Endoplasmic Reticulum Stress Model of Cerebral Ischemia. Brain Sci. 2015, 5, 178–187.
- Boman, K.; Andersson, G.; Wennersten, C.; Nodin, B.; Ahlgren, G.; Jirström, K. Podocalyxin-like and RNA-binding motif protein 3 are prognostic biomarkers in urothelial bladder cancer: A validatory study. Biomark. Res. 2017, 5, 10.
- Boman, K.; Segersten, U.; Ahlgren, G.; Eberhard, J.; Uhlén, M.; Jirström, K.; Malmström, P. Decreased expression of RNA-binding motif protein 3 correlates with tumour progression and poor prognosis in urothelial bladder cancer. BMC Urol. 2013, 13, 17.
- Florianova, L.; Xu, B.; Traboulsi, S.; Elmansi, H.; Tanguay, S.; Aprikian, A.; Kassouf, W.; Brimo, F. Evaluation of RNA-binding motif protein 3 expression in urothelial carcinoma of the bladder: An immunohistochemical study. World J. Surg. Oncol. 2015, 13, 317.
- Bartkova, J.; Rajpert-De Meyts, E.; Skakkebaek, N.; Lukas, J.; Bartek, J. DNA damage response in human testes and testicular germ cell tumours: Biology and implications for therapy. Int. J. Androl. 2007, 30, 282–291, discussion 291.
- Karnevi, E.; Dror, L.; Mardinoglu, A.; Elebro, J.; Heby, M.; Olofsson, S.; Nodin, B.; Eberhard, J.; Gallagher, W.; Uhlén, M.; et al. Translational study reveals a two-faced role of RBM3 in pancreatic cancer and suggests its potential value as a biomarker for improved patient stratification. Oncotarget 2018, 9, 6188–6200.
- Moss, E.; Lee, R.; Ambros, V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 1997, 88, 637–646.
- Zhou, J.; Ng, S.; Chng, W. LIN28/LIN28B: An emerging oncogenic driver in cancer stem cells. Int. J. Biochem. Cell Biol. 2013, 45, 973–978.
- Huang, Y. A mirror of two faces: Lin28 as a master regulator of both miRNA and mRNA. Wiley Interdiscip. Rev. RNA 2012, 3, 483–494.
- Heo, I.; Joo, C.; Cho, J.; Ha, M.; Han, J.; Kim, V. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell 2008, 32, 276–284.
- Lu, L.; Katsaros, D.; Shaverdashvili, K.; Qian, B.; Wu, Y.; de la Longrais, I.; Preti, M.; Menato, G.; Yu, H. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur. J. Cancer 2009, 45, 2212–2218.
- Lei, X.; Xu, J.; Ma, W.; Qiao, C.; Newman, M.; Hammond, S.; Huang, Y. Determinants of mRNA recognition and translation regulation by Lin28. Nucleic Acids Res. 2012, 40, 3574–3584.
- Balzer, E.; Moss, E. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007, 4, 16–25.
- Piskounova, E.; Polytarchou, C.; Thornton, J.; LaPierre, R.; Pothoulakis, C.; Hagan, J.; Iliopoulos, D.; Gregory, R. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 2011, 147, 1066–1079.
- Calin, G.; Sevignani, C.; Dumitru, C.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004.
- Chirshev, E.; Oberg, K.; Ioffe, Y.; Unternaehrer, J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24.
- Chen, Y.; Xie, C.; Zheng, X.; Nie, X.; Wang, Z.; Liu, H.; Zhao, Y. let-7LIN28//PD-L1 Pathway as a Target for Cancer Immunotherapy. Cancer Immunol. Res. 2019, 7, 487–497.
- Ambros, V.; Horvitz, H. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 1984, 226, 409–416.
- Balzeau, J.; Menezes, M.; Cao, S.; Hagan, J. The LIN28/let-7 Pathway in Cancer. Front. Genet. 2017, 8, 31.
- Kang, M.; Lee, K.; Lee, H.; Jeong, C.; Ku, J.; Kim, H.; Kwak, C. Concurrent treatment with simvastatin and NF-κB inhibitor in human castration-resistant prostate cancer cells exerts synergistic anti-cancer effects via control of the NF-κB/LIN28/let-7 miRNA signaling pathway. PLoS ONE 2017, 12, e0184644.
- Büssing, I.; Slack, F.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409.
- Viswanathan, S.; Powers, J.; Einhorn, W.; Hoshida, Y.; Ng, T.; Toffanin, S.; O’Sullivan, M.; Lu, J.; Phillips, L.; Lockhart, V.; et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009, 41, 843–848.
- Rybak, A.; Fuchs, H.; Smirnova, L.; Brandt, C.; Pohl, E.; Nitsch, R.; Wulczyn, F. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008, 10, 987–993.
- Dangi-Garimella, S.; Yun, J.; Eves, E.; Newman, M.; Erkeland, S.; Hammond, S.; Minn, A.; Rosner, M. Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. EMBO J. 2009, 28, 347–358.
- Chang, T.; Zeitels, L.; Hwang, H.; Chivukula, R.; Wentzel, E.; Dews, M.; Jung, J.; Gao, P.; Dang, C.; Beer, M.; et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. USA 2009, 106, 3384–3389.
- Iliopoulos, D.; Hirsch, H.; Struhl, K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009, 139, 693–706.
- Liao, W.; Wang, W.; Chang, W.; Tseng, J. The RNA-binding protein HuR stabilizes cytosolic phospholipase A2α mRNA under interleukin-1β treatment in non-small cell lung cancer A549 Cells. J. Biol. Chem. 2011, 286, 35499–35508.
- Ma, W.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996, 271, 8144–8151.
- Abdelmohsen, K.; Gorospe, M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 2010, 1, 214–229.
- Zarei, M.; Lal, S.; Parker, S.; Nevler, A.; Vaziri-Gohar, A.; Dukleska, K.; Mambelli-Lisboa, N.; Moffat, C.; Blanco, F.; Chand, S.; et al. Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells. Cancer Res. 2017, 77, 4460–4471.
- von Roretz, C.; Di Marco, S.; Mazroui, R.; Gallouzi, I. Turnover of AU-rich-containing mRNAs during stress: A matter of survival. Wiley Interdiscip. Rev. RNA 2011, 2, 336–347.
- Scheiba, R.; de Opakua, A.; Díaz-Quintana, A.; Cruz-Gallardo, I.; Martínez-Cruz, L.; Martínez-Chantar, M.; Blanco, F.; Díaz-Moreno, I. The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets. RNA Biol. 2014, 11, 1250–1261.
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867.
- Han, S.; Tang, Y.; Smith, R. Functional diversity of the hnRNPs: Past, present and perspectives. Biochem. J. 2010, 430, 379–392.
- Piñol-Roma, S.; Choi, Y.; Matunis, M.; Dreyfuss, G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988, 2, 215–227.
- Pawson, T. Protein modules and signalling networks. Nature 1995, 373, 573–580.
- Kiledjian, M.; Dreyfuss, G. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 1992, 11, 2655–2664.
- Cartegni, L.; Maconi, M.; Morandi, E.; Cobianchi, F.; Riva, S.; Biamonti, G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J. Mol. Biol. 1996, 259, 337–348.
- Chen, Y.; Zeng, Y.; Xiao, Z.; Chen, S.; Li, Y.; Zou, J.; Zeng, X. Role of heterogeneous nuclear ribonucleoprotein K in tumor development. J. Cell. Biochem. 2019, 120, 14296–14305.
- Beusch, I.; Barraud, P.; Moursy, A.; Cléry, A.; Allain, F. Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7. eLife 2017, 6, e25736.
- Jensen, K.; Dredge, B.; Stefani, G.; Zhong, R.; Buckanovich, R.; Okano, H.; Yang, Y.; Darnell, R. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron 2000, 25, 359–371.
- Piñol-Roma, S.; Dreyfuss, G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 1992, 355, 730–732.
- Rebane, A.; Aab, A.; Steitz, J. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA 2004, 10, 590–599.
- Allemand, E.; Guil, S.; Myers, M.; Moscat, J.; Cáceres, J.; Krainer, A. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA 2005, 102, 3605–3610.
- Deng, M.; Wang, N.; Li, Z.; Chen, R.; Duan, J.; Peng, Y.; Wu, Z.; Zhang, Z.; Jiang, L.; Zheng, X.; et al. FXR1 can bind with the CFIm25/CFIm68 complex and promote the progression of urothelial carcinoma of the bladder by stabilizing TRAF1 mRNA. Cell Death Dis. 2022, 13, 170.
- Li, P.; Mi, Q.; Yan, S.; Xie, Y.; Cui, Z.; Zhang, S.; Wang, Y.; Gao, H.; Wang, Y.; Li, J.; et al. Characterization of circSCL38A1 as a novel oncogene in bladder cancer via targeting ILF3/TGF-β2 signaling axis. Cell Death Dis. 2023, 14, 59.
- Mancarella, C.; Scotlandi, K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front. Cell Dev. Biol. 2019, 7, 363.
- Huang, X.; Zhang, H.; Guo, X.; Zhu, Z.; Cai, H.; Kong, X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J. Hematol. Oncol. 2018, 11, 88.
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.; Mesquita, A.; Liu, C.; Yuan, C.; et al. Publisher Correction: Recognition of RNA N-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2020, 22, 1288.
- Schneider, T.; Hung, L.; Aziz, M.; Wilmen, A.; Thaum, S.; Wagner, J.; Janowski, R.; Müller, S.; Schreiner, S.; Friedhoff, P.; et al. Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. Nat. Commun. 2019, 10, 2266.
- Wang, P.; Wang, X.; Liu, M.; Zeng, Z.; Lin, C.; Xu, W.; Ma, W.; Wang, J.; Xiang, Q.; Johnston, R.; et al. The Oncogenic Functions of Insulin-like Growth Factor 2 mRNA-Binding Protein 3 in Human Carcinomas. Curr. Pharm. Des. 2020, 26, 3939–3954.
- Lee, D.; Xylinas, E.; Rieken, M.; Khani, F.; Klatte, T.; Wood, C.; Karam, J.; Weizer, A.; Raman, J.; Remzi, M.; et al. Insulin-like growth factor messenger RNA-binding protein 3 expression helps prognostication in patients with upper tract urothelial carcinoma. Eur. Urol. 2014, 66, 379–385.
- Huang, W.; Li, Y.; Zhang, C.; Zha, H.; Zhou, X.; Fu, B.; Guo, J.; Wang, G. IGF2BP3 facilitates cell proliferation and tumorigenesis via modulation of JAK/STAT signalling pathway in human bladder cancer. J. Cell. Mol. Med. 2020, 24, 13949–13960.
- Jia, W.; Yao, Z.; Zhao, J.; Guan, Q.; Gao, L. New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci. 2017, 186, 1–10.
- Berger, C.; Gaume, X.; Bouvet, P. The roles of nucleolin subcellular localization in cancer. Biochimie 2015, 113, 78–85.
- Srivastava, M.; McBride, O.; Fleming, P.; Pollard, H.; Burns, A. Genomic organization and chromosomal localization of the human nucleolin gene. J. Biol. Chem. 1990, 265, 14922–14931.
- Cheng, Y.; Zhao, G.; Zhang, S.; Nigim, F.; Zhou, G.; Yu, Z.; Song, Y.; Chen, Y.; Li, Y. AS1411-Induced Growth Inhibition of Glioma Cells by Up-Regulation of p53 and Down-Regulation of Bcl-2 and Akt1 via Nucleolin. PLoS ONE 2016, 11, e0167094.
- Ginisty, H.; Sicard, H.; Roger, B.; Bouvet, P. Structure and functions of nucleolin. J. Cell Sci. 1999, 112, 761–772.
- Chen, Z.; Xu, X. Roles of nucleolin. Focus on cancer and anti-cancer therapy. Saudi Med. J. 2016, 37, 1312–1318.
- Ugrinova, I.; Monier, K.; Ivaldi, C.; Thiry, M.; Storck, S.; Mongelard, F.; Bouvet, P. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol. Biol. 2007, 8, 66.
- Xu, J.; Lu, S.; Xu, X.; Hu, S.; Li, B.; Qi, R.; Chen, L.; Chang, J. Knocking Down Nucleolin Expression Enhances the Radiosensitivity of Non-Small Cell Lung Cancer by Influencing DNA-PKcs Activity. Asian Pac. J. Cancer Prev. APJCP 2015, 16, 3301–3306.
- Benedetti, E.; Antonosante, A.; d’Angelo, M.; Cristiano, L.; Galzio, R.; Destouches, D.; Florio, T.; Dhez, A.; Astarita, C.; Cinque, B.; et al. Nucleolin antagonist triggers autophagic cell death in human glioblastoma primary cells and decreased in vivo tumor growth in orthotopic brain tumor model. Oncotarget 2015, 6, 42091–42104.
- Subramanian, N.; Srimany, A.; Kanwar, J.; Kanwar, R.; Akilandeswari, B.; Rishi, P.; Khetan, V.; Vasudevan, M.; Pradeep, T.; Krishnakumar, S. Nucleolin-aptamer therapy in retinoblastoma: Molecular changes and mass spectrometry-based imaging. Mol. Ther. Nucleic Acids 2016, 5, e358.
- Shang, Y.; Kakinuma, S.; Nishimura, M.; Kobayashi, Y.; Nagata, K.; Shimada, Y. Interleukin-9 receptor gene is transcriptionally regulated by nucleolin in T-cell lymphoma cells. Mol. Carcinog. 2012, 51, 619–627.
- D’Avino, C.; Palmieri, D.; Braddom, A.; Zanesi, N.; James, C.; Cole, S.; Salvatore, F.; Croce, C.; De Lorenzo, C. A novel fully human anti-NCL immunoRNase for triple-negative breast cancer therapy. Oncotarget 2016, 7, 87016–87030.
- Palmieri, D.; Richmond, T.; Piovan, C.; Sheetz, T.; Zanesi, N.; Troise, F.; James, C.; Wernicke, D.; Nyei, F.; Gordon, T.; et al. Human anti-nucleolin recombinant immunoagent for cancer therapy. Proc. Natl. Acad. Sci. USA 2015, 112, 9418–9423.
- Gilles, M.; Maione, F.; Cossutta, M.; Carpentier, G.; Caruana, L.; Di Maria, S.; Houppe, C.; Destouches, D.; Shchors, K.; Prochasson, C.; et al. Nucleolin Targeting Impairs the Progression of Pancreatic Cancer and Promotes the Normalization of Tumor Vasculature. Cancer Res. 2016, 76, 7181–7193.
- Fogal, V.; Sugahara, K.; Ruoslahti, E.; Christian, S. Cell surface nucleolin antagonist causes endothelial cell apoptosis and normalization of tumor vasculature. Angiogenesis 2009, 12, 91–100.
- Ding, Y.; Song, N.; Liu, C.; He, T.; Zhuo, W.; He, X.; Chen, Y.; Song, X.; Fu, Y.; Luo, Y. Heat shock cognate 70 regulates the translocation and angiogenic function of nucleolin. Arterioscler. Thromb. Vasc. Biol. 2012, 32, e126–e134.
- Qian, B.; Yao, Y.; Liu, Y.; Yan, M.; Huang, Y.; Chen, Y. Nucleolin identified by comparative mass-spectra analysis is a potential marker for invasive progression of hepatocellular carcinoma. Mol. Med. Rep. 2014, 10, 1489–1494.
- Li, Y.; Tang, Y.; Ye, L.; Liu, B.; Liu, K.; Chen, J.; Xue, Q. Establishment of a hepatocellular carcinoma cell line with unique metastatic characteristics through in vivo selection and screening for metastasis-related genes through cDNA microarray. J. Cancer Res. Clin. Oncol. 2003, 129, 43–51.
- Choi, W.; Yang, Y.; Xu, Y.; An, J. Targeting chemokine receptor CXCR4 for treatment of HIV-1 infection, tumor progression, and metastasis. Curr. Top. Med. Chem. 2014, 14, 1574–1589.
- Niu, H.; Yang, X.; Xu, Z.; Du, T.; Wang, R. Cell surface nucleolin interacts with CXCR4 receptor via the 212 c-terminal portion. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 1099–1104.
- Yu, Y.; Jin, H.; Xu, J.; Gu, J.; Li, X.; Xie, Q.; Huang, H.; Li, J.; Tian, Z.; Jiang, G.; et al. XIAP overexpression promotes bladder cancer invasion in vitro and lung metastasis in vivo via enhancing nucleolin-mediated Rho-GDIβ mRNA stability. Int. J. Cancer 2018, 142, 2040–2055.
- Ren, S.; Zhang, N.; Shen, L.; Lu, Y.; Chang, Y.; Lin, Z.; Sun, N.; Zhang, Y.; Xu, J.; Huang, H.; et al. Lnc00892 competes with c-Jun to block NCL transcription, reducing the stability of RhoA/RhoC mRNA and impairing bladder cancer invasion. Oncogene 2021, 40, 6579–6589.
- Jin, H.; Yu, Y.; Hu, Y.; Lu, C.; Li, J.; Gu, J.; Zhang, L.; Huang, H.; Zhang, D.; Wu, X.; et al. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget 2015, 6, 522–536.
- Daizumoto, K.; Yoshimaru, T.; Matsushita, Y.; Fukawa, T.; Uehara, H.; Ono, M.; Komatsu, M.; Kanayama, H.; Katagiri, T. A DDX31/Mutant-p53/EGFR Axis Promotes Multistep Progression of Muscle-Invasive Bladder Cancer. Cancer Res. 2018, 78, 2233–2247.
- Lubet, R.; Lu, Y.; Bode, A.; You, M.; Verney, Z.; Steele, V.; Townsend, R.; Juliana, M.; Grubbs, C. Efficacy of the EGFr inhibitor Iressa on development of chemically-induced urinary bladder cancers: Dose dependency and modulation of biomarkers. Oncol. Rep. 2011, 25, 1389–1397.
- Vernet, C.; Artzt, K. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. TIG 1997, 13, 479–484.
- Artzt, K.; Wu, J. STAR trek: An introduction to STAR family proteins and review of quaking (QKI). Adv. Exp. Med. Biol. 2010, 693, 1–24.
- Teplova, M.; Hafner, M.; Teplov, D.; Essig, K.; Tuschl, T.; Patel, D. Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev. 2013, 27, 928–940.
- Darbelli, L.; Richard, S. Emerging functions of the Quaking RNA-binding proteins and link to human diseases. Wiley Interdiscip. Rev. RNA 2016, 7, 399–412.
- Farnsworth, B.; Peuckert, C.; Zimmermann, B.; Jazin, E.; Kettunen, P.; Emilsson, L. Gene Expression of Quaking in Sporadic Alzheimer’s Disease Patients is Both Upregulated and Related to Expression Levels of Genes Involved in Amyloid Plaque and Neurofibrillary Tangle Formation. J. Alzheimer’s Dis. 2016, 53, 209–219.
- Galarneau, A.; Richard, S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat. Struct. Mol. Biol. 2005, 12, 691–698.
- Ryder, S.; Frater, L.; Abramovitz, D.; Goodwin, E.; Williamson, J. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 2004, 11, 20–28.
- Zhang, R.; Yang, J.; Peng, L.; Zheng, L.; Xie, P.; Wang, M.; Cao, Y.; Zhang, Z.; Zhou, F.; Qian, C.; et al. RNA-binding protein QKI-5 inhibits the proliferation of clear cell renal cell carcinoma via post-transcriptional stabilization of RASA1 mRNA. Cell Cycle 2016, 15, 3094–3104.
- Novikov, L.; Park, J.; Chen, H.; Klerman, H.; Jalloh, A.; Gamble, M. QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 2011, 31, 4244–4255.
- Bian, Y.; Wang, L.; Lu, H.; Yang, G.; Zhang, Z.; Fu, H.; Lu, X.; Wei, M.; Sun, J.; Zhao, Q.; et al. Downregulation of tumor suppressor QKI in gastric cancer and its implication in cancer prognosis. Biochem. Biophys. Res. Commun. 2012, 422, 187–193.
- Yang, G.; Fu, H.; Zhang, J.; Lu, X.; Yu, F.; Jin, L.; Bai, L.; Huang, B.; Shen, L.; Feng, Y.; et al. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology 2010, 138, 231–240.e5.
- Zhao, Y.; Zhang, G.; Wei, M.; Lu, X.; Fu, H.; Feng, F.; Wang, S.; Lu, W.; Wu, N.; Lu, Z.; et al. The tumor suppressing effects of QKI-5 in prostate cancer: A novel diagnostic and prognostic protein. Cancer Biol. Ther. 2014, 15, 108–118.
- Shi, F.; Deng, Z.; Zhou, Z.; Jiang, C.; Zhao, R.; Sun, F.; Cui, D.; Bei, X.; Yang, B.; Sun, Q.; et al. QKI-6 inhibits bladder cancer malignant behaviours through down-regulating E2F3 and NF-κB signalling. J. Cell. Mol. Med. 2019, 23, 6578–6594.
- Wei, X.; Wang, B.; Wang, Q.; Yang, X.; Yang, Y.; Fang, Z.; Yi, C.; Shi, L.; Fan, X.; Tao, J.; et al. MiR-362-5p, Which Is Regulated by Long Non-Coding RNA MBNL1-AS1, Promotes the Cell Proliferation and Tumor Growth of Bladder Cancer by Targeting QKI. Front. Pharmacol. 2020, 11, 164.
- LeBleu, V.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Model. Mech. 2018, 11, dmm029447.
- Quail, D.; Joyce, J. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437.
- Kharaishvili, G.; Simkova, D.; Bouchalova, K.; Gachechiladze, M.; Narsia, N.; Bouchal, J. The role of cancer-associated fibroblasts, solid stress and other microenvironmental factors in tumor progression and therapy resistance. Cancer Cell Int. 2014, 14, 41.
- Zhou, Z.; Cui, D.; Sun, M.; Huang, J.; Deng, Z.; Han, B.; Sun, X.; Xia, S.; Sun, F.; Shi, F. CAFs-derived MFAP5 promotes bladder cancer malignant behavior through NOTCH2/HEY1 signaling. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 7970–7988.
More
