Several studies suggest the importance of preserving the micro-ecosystem of commensal, symbiotic and pathogenic microbes of the respiratory system, and, more recently, its relationship with the intestinal microbiome.
However, recent studies investigate how it leads to the maintenance of human health, also in relation to some factors such as the effect of age, diet and others, making its composition and stability better understood.
- human microbiome
- respiratory microbiome
- clinical microbiology
- immune modulation
- dysbiosis
- respiratory diseases
- translational research
- SARS-CoV-2
1. Introduction
Figure 1. Main bacteria genera and microbiome species of an adult in the upper respiratory tract.
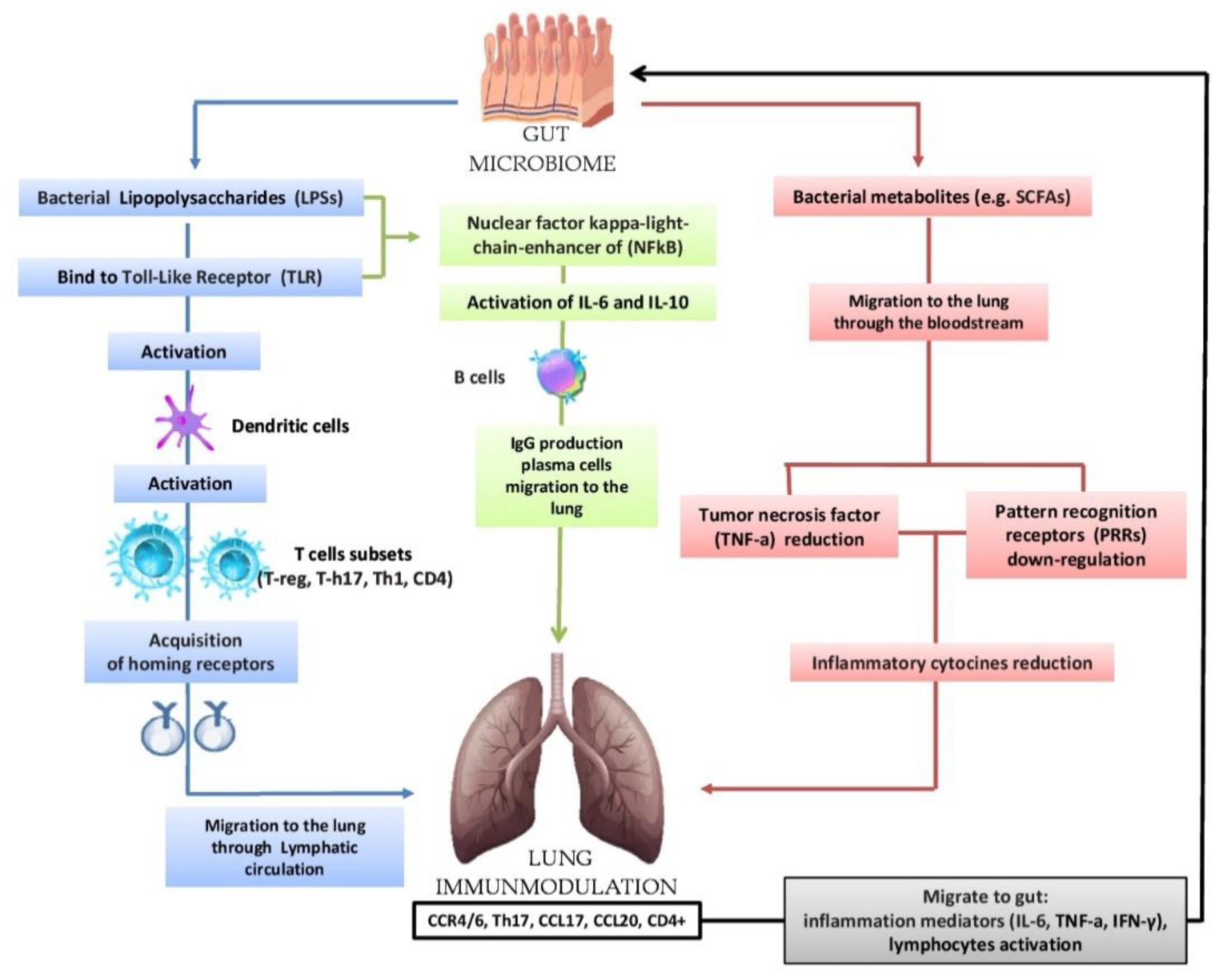
Figure 2.
Ne Bidirectional hypothesis that the intestinal microbiota can modulate the immunological activity of the lung: lipopolysaccharides (LPSs) are able to bind to the Toll-Like Receptor (TLR) present on the intestinal mucosa causing the activation of dendritic cellsdata in the current scientific literature show that the composition of the respiratory system microbiome differs in health and disease conditions an that favor the activation of various T cells (T-reg, T-h17, Th-1, CD4); subsequently, there is the acquisition of horning receptors (CCR6, CCR9, CCR4, a4b7) and migration the microbial community as a collective entity can contribut to the lung through the circulatory lymphatic flow (CCR4/6Treg, CCR4/6Th17, CCR4/6Th1, CCR4/6CD4 +). It also activates (IL-18, INF-y, TNF-a, TGF-b, IL-4, IL-1), which will pass into the circulation (INF-y, TNF-a, IL-6) pathophysiological processes associated with chronic airway diseas. The nuclear factor kappa-light chain enhancer of (NFkB) activates IL-6, IL-10 and the production of IgA and IgG plasma cell b cells and the migration of IgG respiratory microbiome is less studied than that of other areas, but it is believed to contributeto the lung. In the lung, we have the increase in CCL17, CCL20 and the presence of CCR4/6, CD4 +, CCR4/6 and Th17. The migration of bacterial metabolites (e.g., short-chain fatty acids (SCFAs) to the lung through the bloodstream results in the down-regulation of pattern recognition receptors (PRR) with a consequent reduction in the production of inflammatory cytokines (IL-1, IL-12, IL-18), tumor necrosis factor alpha (TNF-α), interferon gamma (IFNγ) and granulocyte–macrophage colony stimulating factor (GM-CSF). In turn, the lungs send inflammation mediators and lymphocytes to the gut in case host’s local immune education and to the development of respiratory diseases, including allergies, asthma and others. In this review, was highlighted the current clinical microbiology knowledge about the microbiota and the various lung diseases relationships, previously only indirectly related to microbial pathogenesis, and the microbiota–pathogenesis relationship of lung infection, among the main cause of disease.
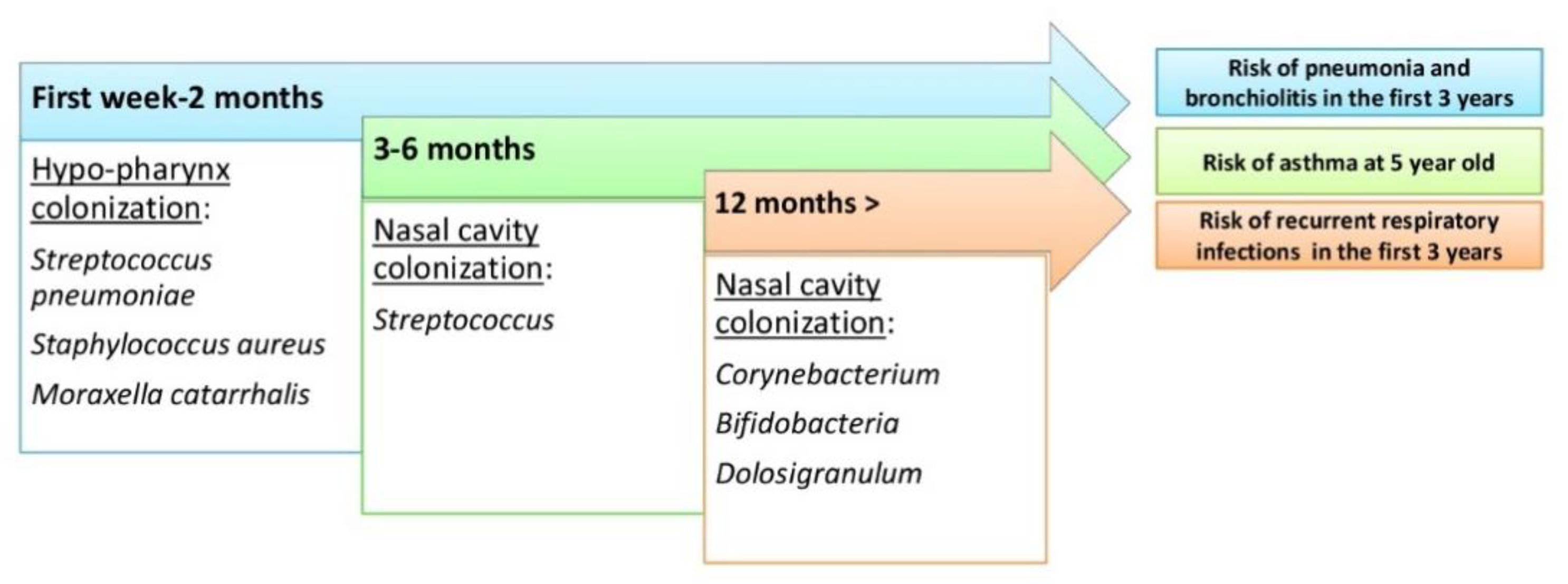
Figure 3.es The evolution of the upper respiratory tract microbiome during human life and risk of respiratory tractin order to prevent and help in a targeted treatment of various lun diseases..
2. The Upper Respiratory Tract Microbiome (Airways Colonization and Evolution during Life)
The upper respiratory system is anatomically related to an interconnected cavity system that includes the nostrils, rhino-pharynx, and oropharynx, and communicates with the larynx and the middle cavity of the ear through the Eustachian tube. The mucous surfaces of these areas are colonized by a wide range of bacteria belonging to the genera Firmicutes, Actinobacteria, Bacteroidetes, Proteobacteria and Fusobacteria[1][4][7]. The main functions of the upper respiratory system are to filter, heat and humidify the air that passes through it before it reaches the lungs. The individual anatomical regions have their own special characteristics (humidity, temperature, relative oxygen concentration, type of epithelial cells, etc.) They create their microenvironment with the result that there are differences in the microbiome at lower taxonomic levels of the microorganisms that colonize each region. The composition of the microbial community is influenced by environmental factors and the interactions of microbes and their host immune system. Normal microbiota prevents the formation of pathogenic microorganisms. The pathogen competes for attachment sites, for nutrients and is difficult to install to multiply and cause disease. Competition between germs is also a factor shaping the microbiome[8][9][10][11]. Colonization of the upper respiratory tract begins at birth and the association of the original microbiome with health established throughout a person’s life with three main modalities: (a) method of delivery (caesarean or normal); (b) environment (habitat, diet etc.); and (c) antibiotics. The rhino-pharynx and oropharynx microbiome at birth and infancy affected by the individual’s environmental exposures, including breastfeeding. Environment habitat has an important role in the develop of the immune system of the lung and the exposure in the first months of life to certain bacteria directs the immunological activity of the child. Recent studies have highlighted how the integrity of the composition and correct maturation of the microbiota in the first period of life can influence the prevention of certain lung diseases or can, in the event of its alteration, cause different pathological states[1][14][15][16][17]. In the first period of life, the signals encoded by the microbiome, both intestinal and pulmonary (Gut/Lung axis), are essential to target the maturation of the cells of the epithelium of airways and affect the maturation of the immune system. However, when bacterial colonization took place in the first year of their age, there seemed to be no association with hissing (wheezing) breath. At 1.5 months of age, five groups predominate: Streptococcus, Moraxella, Staphylococcus, Corynebacterium or Corynebacterium/Dolosigranulum (colonization by S. pneumoniae frequently occurs in children and is asymptomatic and this means that colonization is short and progresses towards infection). The rhino-pharyngeal microbiota of the elderly seems to undergo profound changes. However, it is unclear how these changes affect the composition and maintenance of the microbiome in the upper respiratory tract[1][18][19][20].
2.1. Nostrils
The area of the nostril does not differ much in the characteristics between children and adults. However, children have an abundance of Streptococcaceae, Moraxellaceae and Neisseriaceae families, which have not been confirmed as a mere dispersion from the nearby rhino-pharynx where they predominate or are the result of substantial differentiation of the microbial community from that of adults. In addition, age-related local immunity affects the composition of the microbiome. Antimicrobial peptides, local immune cells such as neutrophils and Natural killer cells (NK), are the first defense against microorganisms[1][5][10][11]. The nostril microbiome is enriched mainly with members of the genera Actinobacteria (Corynebacterium and Propionibacterium spp.) and Firmicutes (Streptococcus species in children and Staphylococcus species in adults). In small amounts, there are anaerobes belonging to the genus Bacteroidetes. As for Proteobacteria, their number varies greatly with studies that report a high abundance of Moraxellaceae in children and members of the class Gammaproteobacteria in healthy adults [1][8][18]. The epithelium of the nostrils contains glands that secrete sebum, and this is related to its selective enrichment with lipophilic bacteria such as Propionibacterium spp., able to metabolize sebum lipids into short-chain fatty acids. The pH decreases and promotes the growth of Corynebacterium and Staphylococcus coagulase. Moreover, the nostrils are rich in oxygen and humidity of contributes to the growth of Staphylococcus aureus and Corynebacterium. Therefore, the simultaneous presence of Propionibacterium and Staphylococcus spp. can be supported by different characteristics of the local environment. Their coexistence can be supported by the production of coproporphyrin III by Propionibacterium spp., which promotes the formation of the Staphylococcus aureus biofilm[4][10][18].
2.2. Rhinopharynx
The rhino-pharyngeal microbiome ripples from the beginning of life onwards. Initially, there is a predominance of species belonging to the genera Moraxella, Corynebacterium, Dolosigranulum, Streptococcus or Staphylococcus spp. and possibly related to the mode of delivery (during caesarean section microbiome: Staphylococcus and Corynebacterium spp. and the microbiome during vaginal delivery: Staphylococcus, Streptococcus and Dolosigranulum spp.), and the type of diet. Later, in adults, there is a clear absence of Moraxella. The species that colonize the rhino-pharynx overlap with species from nearby anatomical areas such as the oropharynx (Streptococcus spp) and the anterior roots (mainly aerobic Gram-positive such as Staphylococcus, Dolosigranulum, Corynebacterium) [7][10][11]. The Gram-negative anaerobes are the same found mainly in the oropharynx and oral cavity, such as Prevotella and Veillonella spp mainly in the rhino-pharynx of young children. The rhino-pharyngeal microbiome plays an important and beneficial role in maintaining the balance of related species, preventing the growth of pathogens and host immunity[18][19][20][21].
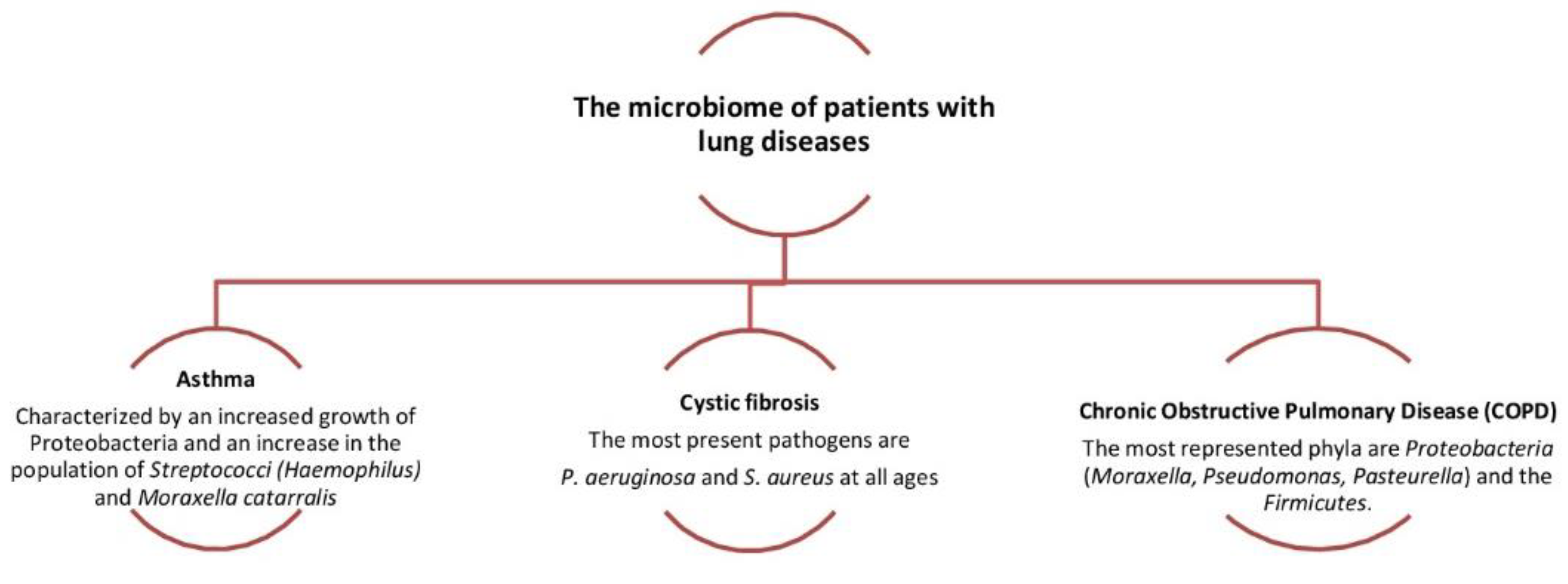
Figure 4. Dysbiosis related to some lung diseases. The relative abundance in
2.3. Oropharynx
The oropharynx, due to its location, is exposed to a wide variety of microorganisms of endogenous and exogenous origin. It is anatomically linked to the oral cavity, the rhino-pharynx, the larynx, the lower respiratory tract, and the gastrointestinal tract. The germs of the pharyngeal oral community can be transmitted to the lower respiratory tract in healthy and inhaled patients. In the healthy adult, the rhino-pharynx is shown to colonize the genera Streptococcus, Haemophilus and Neisseria spp. and Gram-negative anaerobic species Veillonella,Prreoteoella, Leptotrichia and Fusbacteriium. Several pathogens of the genus Streptococcus are found in the pharynx, such as S. pneumona
is associated with the increased severity of chronic obstructive pulmonary disease (COPD).
ae, S. pyogenes (which can cause serious disorders that are not limited to the pharynx, and can also cause septic shock) and S. agalactiae (Figure 1) [1][10][18][22][23].
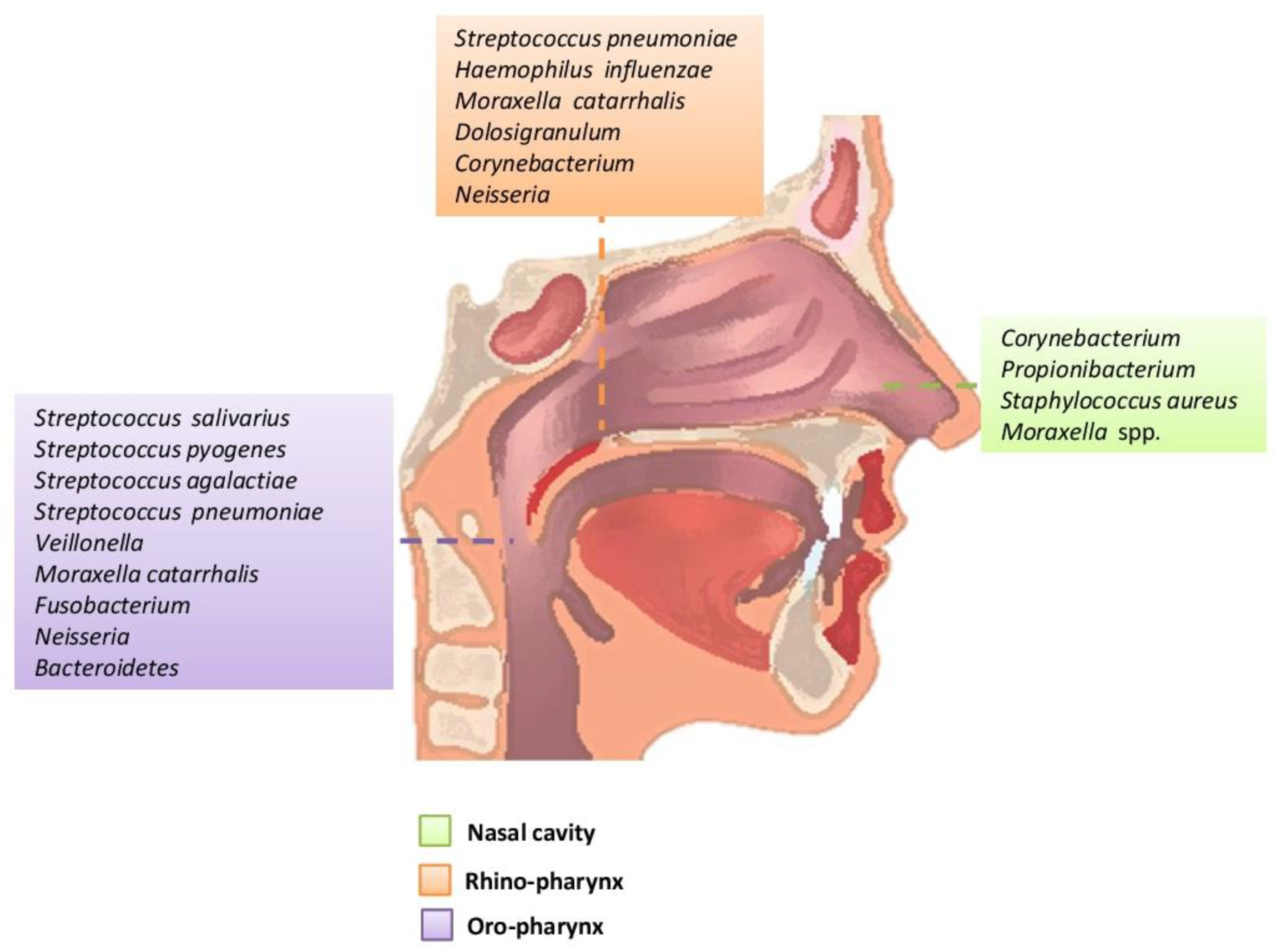
Figure 1. Main bacteria genera and microbiome species of an adult in the upper respiratory tract.
[1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23
References
- Finlay, B.B.; Finlay, J.M. The Whole-Body Microbiome: How to Harness Microbes―Inside and Out―for Lifelong Health Hardcover, 1st ed.; The Experiment: New York, NY, USA, 2019; pp. 238–260.
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810.
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. The Human Microbiome Project Consortium. Structure function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214.
- Sahin-Yilmaz, A.; Naclerio, R.M. Anatomy and physiology of the upper airway. Proc. Am. Thorac. Soc. 2011, 8, 31–39.
- Rogers, G.B.; Shaw, D.; Marsh, R.L.; Carroll, M.P.; Serisier, D.J.; Bruce, K.D. Respiratory microbiota: Addressing clinical questions, informing clinical practice. Thorax 2015, 70, 74–81.
- Mitchell, A.B.; Oliver, B.G.; Glanville, A.R. Translational aspects of the human respiratory virome. Am. J. Respir. Crit. Care Med. 2016, 194, 1458–1464.
- Campanella, V.; Syed, J.; Santacroce, L.; Saini, R.; Ballini, A.; Inchingolo, F. Oral probiotics influence oral and respiratory tract infections in pediatric population: A randomized double-blinded placebo-controlled pilot study. Eur Rev Med Pharmacol Sci. 2018, 22, 8034–8041.
- Biesbroek, G.; Tsivtsivadze, E.; Sanders, E.A.; Montijn, R.; Veenhoven, R.H.; Keijser, B.J.; Bogaert, D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014, 190, 1283–1292.
- Huffnagle, G.; Dickson, R.; Lukacs, N. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal Immunol. 2017, 10, 299–306.
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 17, 87.
- Schenck, L.P.; Surette, M.G.; Bowdish, D.M.E. Composition and immunological significance of the upper respiratory tract microbiota. FEBS Lett. 2016, 21, 3705–3720.
- Ranucci, G.; Buccigrossi, V.; de Freitas, M.B.; Guarino, A.; Giannattasio, A. Early-Life Intestine Microbiota and Lung Health in Children. J. Immunol. Res. 2017, 2017.
- Zhang, D.; Li, S.; Wang, N.; Tan, H.Y.; Zhang, Z.; Feng, Y. The Cross-Talk between Gut Microbiota and Lungs in Common Lung Diseases. Front. Microbiol. 2020, 25, 301.
- Gallacher, D.J.; Kotecha, S. Respiratory Microbiome of New-Born Infants. Front. Pediatr. 2016, 23, 4–10.
- Cicinelli, E.; Ballini, A.; Marinaccio, M.; Poliseno, A.; Coscia, M.F.; Monno, R.; De Vito, D. Microbiological findings in endometrial specimen: Our experience. Arch. Gynecol. Obstet. 2012, 285, 1325–1329.
- DiGiulio, D.B. Diversity of microbes in amniotic fluid. Semin. Fetal Neonatal Med. 2012, 17, 2–11.
- Krzych-Fałta, E.; Furmańczyk, K.; Lisiecka-Biełanowicz, M.; Sybilski, A.; Tomaszewska, A.; Raciborski, F.; Wojas, O.; Walkiewicz, A.; Samel-Kowalik, P.; Samoliński, B. The effect of selected risk factors, including the mode of delivery, on the development of allergic rhinitis and bronchial asthma. Adv. Dermatol. Allergol. 2018, 35, 267–273.
- David, W.; Cleary, S.C. Clarke the nasopharyngeal microbiome Emerging Topics in Life. Sciences 2017, 1, 297–312.
- Bosch, A.A.T.M.; Levin, E.; van Houten, M.A.; Hasrat, R.; Kalkman, G.; Biesbroek, G.; de Steenhuijsen Piters, W.A.A.; de Groot, P.C.M.; Pernet, P.; Keijser, B.J.F.; et al. Development of Upper Respiratory Tract Microbiota in Infancy is Affected by Mode of Delivery. EBioMedicine 2016, 9, 336–345.
- Ballini, A.; Cantore, S.; Farronato, D.; Cirulli, N.; Inchingolo, F.; Papa, F.; Malcangi, G.; Inchingolo, A.D.; Dipalma, G.; Sardaro, N.; et al. Periodontal disease and bone pathogenesis: The crosstalk between cytokines and porphyromonas gingivalis. J. Biol. Regul. Homeost. Agents 2015, 29, 273–281.
- Gnoni, A.; De Nitto, E.; Scacco, S.; Santacroce, L.; Palese, L.L. A New Look at the Structures of Old Sepsis Actors by Exploratory Data Analysis Tools. Antibiotics 2019, 8, 225.
- Di Serio, F.; Lovero, R.; D’Agostino, D.; Nisi, L.; Miragliotta, G.; Contino, R.; Man, A.; Ciccone, M.M.; Santacroce, L. Evaluation of procalcitonin, Vitamin D and C-reactive protein levels in septic patients with positive emocoltures. Our preliminary experience. Acta Med. Mediterr. 2016, 32, 1911–1914.
- Ballini, A.; Dipalma, G.; Isacco, C.G.; Boccellino, M.; Di Domenico, M.; Santacroce, L.; Nguyễn, K.C.D.; Scacco, S.; Calvani, M.; Boddi, A.; et al. Oral Microbiota and Immune System Crosstalk: A Translational Research. Biology 2020, 9, 131.
