Molecular dynamics (MD) simulations are powerful theoretical methods that can reveal biomolecular properties, such as structure, fluctuations, and ligand binding, at the atomic level. All-atom MD simulations elucidated a difference in the dynamic properties of RNA-dependent RNA polymerases (RdRps) in severe acute respiratory syndrom coronavirus 2 (SARS-CoV-2) and SARS-CoV, which may cause activity differences of these RdRps. RdRp is also a drug target for Coronavirus disease 2019. Nucleotide analogs, such as remdesivir and favipiravir, are considered to be taken up by RdRp and inhibit RNA replication. The recognition mechanism of RdRp for these drug molecules and adenosine triphosphate (ATP) was revealed by MD simulations at the atomic detail. In addition, various simulation studies on the complexes of SARS-CoV-2 RdRp with several nucleotide analogs are also presented.
- molecular dynamics simulation
- RNA-dependent RNA polymerase
- SARS-CoV-2
- nucleotide analogs
- RNA replication inhibition
1. Introduction
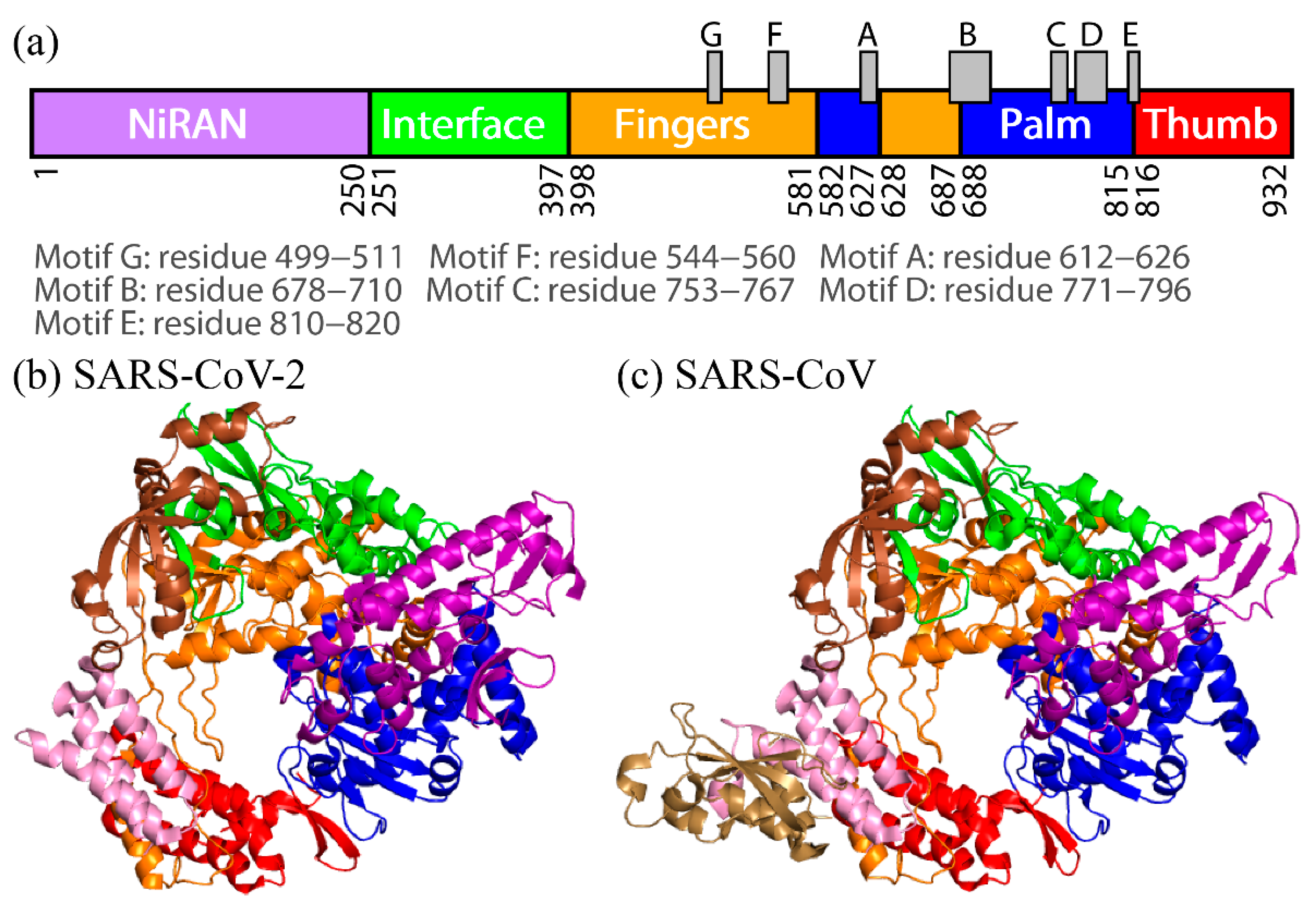
2. Difference in Dynamic Properties of SARS-CoV and SARS-CoV-2 RdRps
To observe the tertiary-structure difference between nsp12s of SARS-CoV-2 and SARS-CoV, the average distances between Cα atoms in nsp12s were calculated as shown in Figure 3a and 3b. WResearchers can see that the two systems have the following in common: the NiRAN and palm domains are spatially close to each other, and the interface and fingers domains are close to each other. To clarify the differences in the average distances between nsp12s of SARS-CoV-2 and SARS-CoV, Itoh et al. calculated the ratio of the difference (Figure 3c). The differences between nsp12s of SARS-CoV-2 and SARS-CoV are observed in the region indicated by the brown square. Blue lines (or blue meshes) are observed in residues around 430, 520, 560, 620, 690, 760, and 800. These results mean that the distances between all motifs of nsp12 in SARS-CoV are shorter than those of nsp12 in SARS-CoV-2. In particular, the distance between motifs F and G for SARS-CoV is up to 63% shorter than that for SARS-CoV-2.
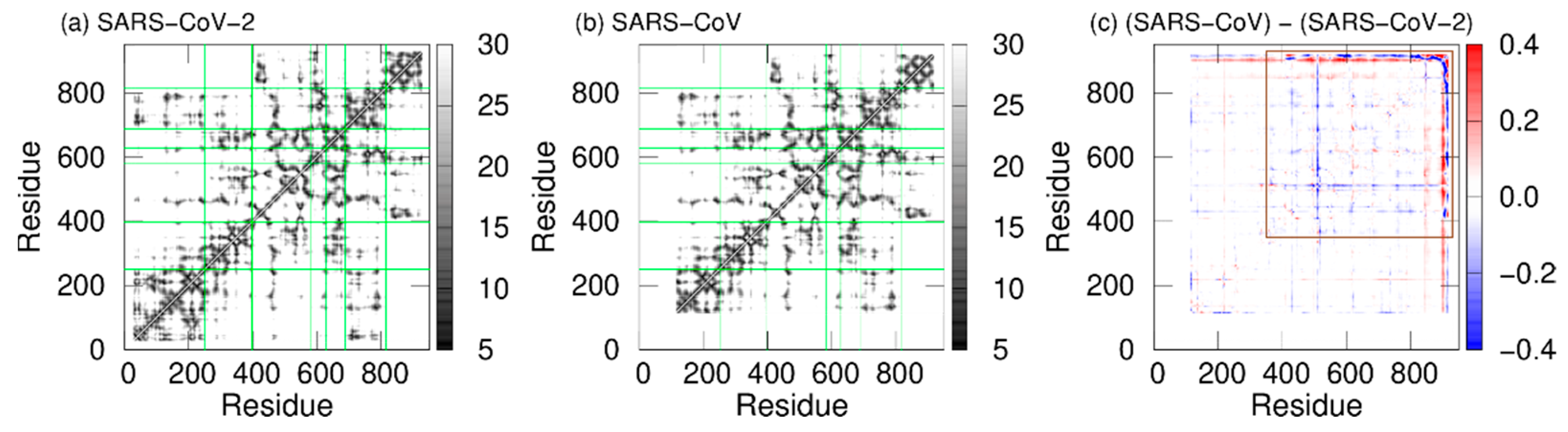
Figure 3. The average distances between Cα atoms of nsp12 for (a) SARS-CoV-2 and (b) SARS-CoV. The borders between the domains in nsp12 are indicated by the green lines. (c) The ratios of the differences between the average distances for SARS-CoV nsp12 and those for SARS-CoV-2 nsp12. The brown square shows residues that have large differences. Reproduced with permission from Ref. [17].
In addition, dynamic cross-correlation (DCC) was calculated to investigate the correlation between domain motions. DCCs of SARS-CoV-2 nsp12 and SARS-CoV nsp12 are presented in Figure 4a and 4b. Here, red and blue indicate positive and negative correlations, respectively. The fact that there is a positive (negative) correlation between two residues indicates that the motions of these residues are in the same (opposite) direction. In both systems, positive correlations are found between most residues within the same domains. However, there are both positive and negative correlations in the interface domain of SARS-CoV nsp12. The boundary between these correlations is residue 330. Residues before and after residue 330 in the interface domain are positively correlated with the NiRAN domain and fingers domain, respectively. Figure 4c shows the differences in DCCs between SARS-CoV-2 and SARS-CoV nsp12s. As shown by the region surrounded by the brown lines, the differences are larger in the NiRAN and interface domains. These domains before residue 330 have a strong negative correlation with the fingers domain in SARS-CoV nsp12. That is, the regions before residue 330 move cooperatively with the fingers domain, moving closer and further away from each other.
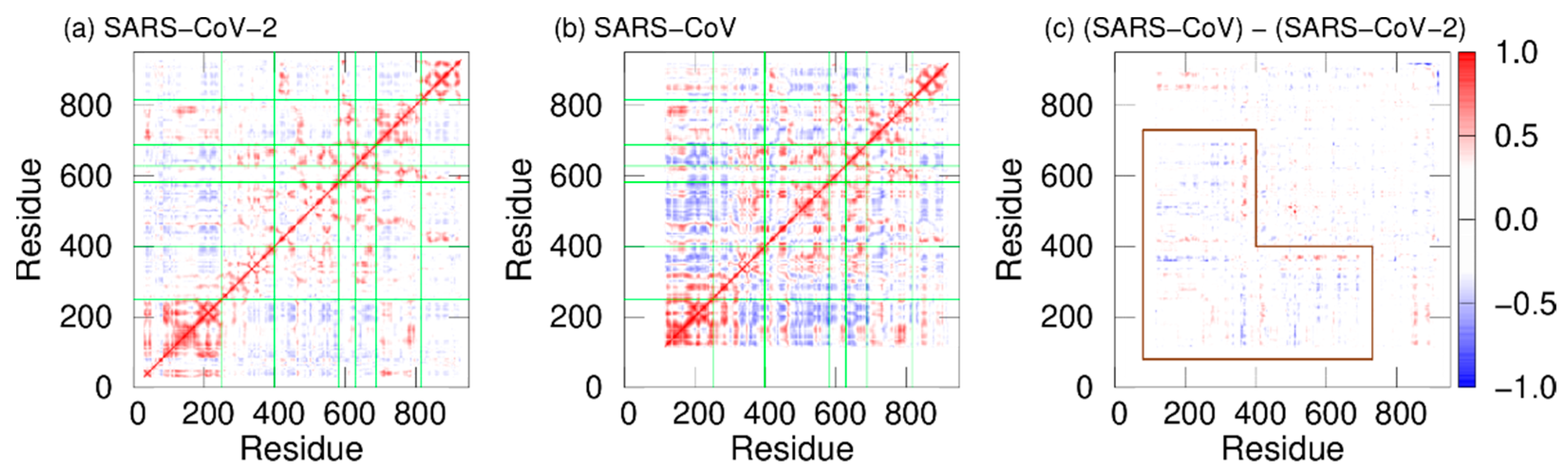
Figure 4. DCCs of nsp12 for (a) SARS-CoV-2 and (b) SARS-CoV. The borders between the domains in nsp12 are indicated by the green lines. (c) Differences between DCCs for SARS-CoV nsp12 and those for SARS-CoV-2 nsp12. The region surrounded by the brown lines means residues with large differences. Reproduced with permission from Ref. [17].
As shown in Figure 3c, the distances between all motifs in SARS-CoV nsp12 are shorter compared to SARS-CoV-2 nsp12. This may enhance the RdRp activity of SARS-CoV. Furthermore, in SARS-CoV nsp12, the NiRAN and fingers domains move cooperatively toward and away from each other; because the removal of the NiRAN domain reduces the RdRp activity [33], the NiRAN domain is important for the RdRp activities. The cooperative movement of the NiRAN domain with the core (fingers) domain of RdRp may also enhance the activity of RdRp.
3. “Bucket Brigade” in RdRp Ligand Recognition
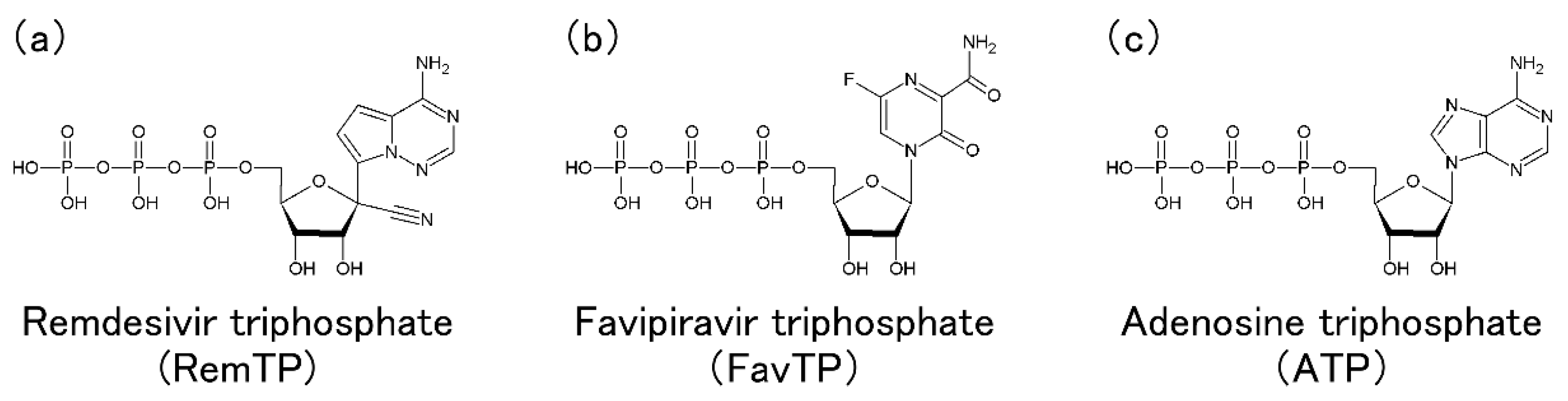
In this section, Tanimoto et al.'s MD simulations of RdRp with RemTP, FavTP, or ATP to clarify how RdRp recognizes the drugs and NTPs are presented [20].
As a result of the MD simulations, the ligand recognition process by RdRp was observed in all three systems of RemTP, FavTP, and ATP. First, the ligand recognition probability was calculated, as listed in Table 1. RemTP shows the highest probability, FavTP shows the second-highest probability, followed by ATP, although within the statistical errors. These results are in qualitative agreement with previous experimental studies [11][34]. In addition, MD simulations of the RdRp-RemTP complex using the free energy perturbation (FEP) method showed that RemTP is bound more strongly to RdRp than ATP [35], which is also consistent with the present results.
Table 1. The number of MD simulations in which RdRp recognized the ligands. Ligand recognition probability is also listed. Reproduced with permission from Ref. [20].
The number of MD simulations in which RdRp recognized the ligands. Ligand recognition probability is also listed.
|
Ligand |
Ligand Recognition/Total |
Ligand Recognition Probability |
|
RemTP |
12/50 |
0.24 ± 0.07 |
|
FavTP |
9/50 |
0.18 ± 0.06 |
|
ATP |
7/50 |
0.14 ± 0.06 |
Next, to understand the mechanism of the ligand recognition by RdRp, the trajectories of the recognized ligands were examined. As a result, an interesting path was observed in which the lysine residues of RdRp carry ligands to the binding site like a “bucket brigade,” as shown in Figure 5. In this path, the phosphate groups of the ligands contacted LYS2 and LYS43 of nsp7 and LYS551, LYS621, and LYS798 of nsp12. Because nsp12 and nsp7 correspond to chain A and chain C, respectively, in the original cryo-EM structure, the residues are expressed here as “chain label + residue number + residue name”. These lysine residues have a positive charge, of which C2LYS, C43LYS, and A551LYS are in a line toward the binding site. In this process of ligand transportation, the phosphate groups of RemTP first interact with the side chain of C2LYS (state 1 (S1), Figure 5b). C2LYS passes RemTP to C43LYS, which is spatially close (state 2 (S2), Figure 5c). C43LYS then passes RemTP to A551LYS (state 3 (S3), Figure 5d). RemTP finally reaches the binding site (state 4 (S4), Figure 5e). The ligand also interacts electrically with A621LYS and A798LYS at the binding site. A similar process was also observed in the FavTP and ATP systems.
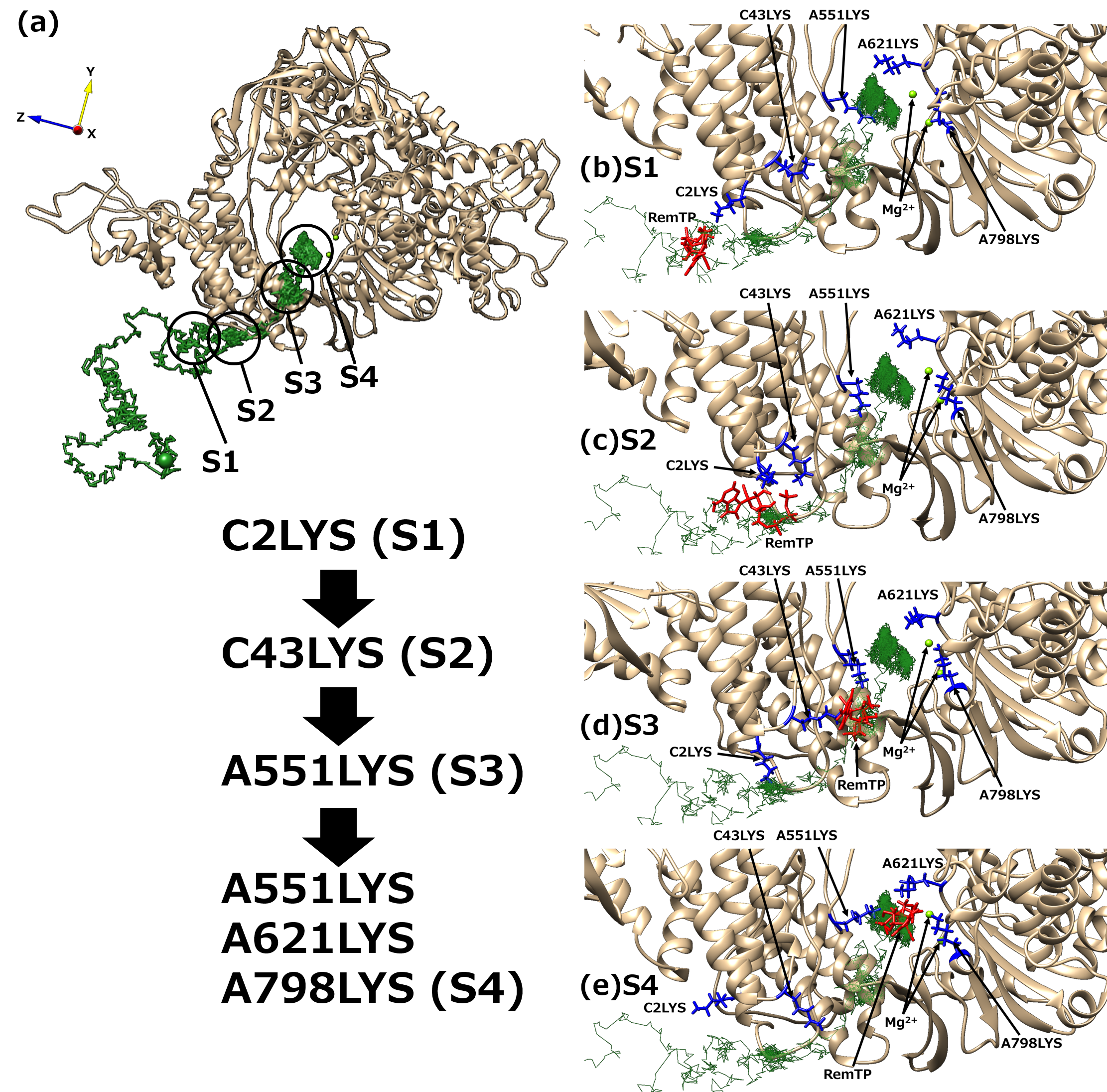
Figure 5. (a) “Bucket brigade” trajectory of RemTP recognized by RdRp. The black circles mean the positions at which RemTP has contact with RdRp residues. (b–e) Typical snapshot at each state (S1–S4). In (b–e), the lysine residues that contributed to the ligand recognition and RemTP are expressed as blue and red stick models, respectively. Reproduced with permission from Ref. [20].
These positively charged residues have been reported to be favorable for the NTP recognition [10][11]. Furthermore, the lysine residues, which contribute to the bucket-brigade ligand transportation, are highly conserved in RdRp of SARS-CoV [12]. Therefore, it is expected that for both SARS-CoV-2 and SARS-CoV RdRps, these linearly arranged lysine residues carry NTPs to the binding site, thereby enhancing the NTP recognition ability of RdRp.
4. Molecular Simulation Studies on Inhibition Mechanisms of Nucleotide Analogs against the SARS-CoV-2 RdRp Function
4.1. SARS-CoV-2 RdRp with Remdesivir in the Triphosphate Form
It is important to understand the mechanism by which RemTP is bound to RdRp and inhibits the RNA replications. Zhang et al. examined how remdesivir integrated into the nascent RNA strand (N-Rem) inhibited RdRp from adding nucleotides to the strand [36]. They revealed that N-Rem led to a delayed chain termination, where the translocation of the nascent RNA strand is terminated once three nucleotides were added after the RemTP incorporation. It was clarified that the forward translocation of the nascent RNA strand was impeded by the electrostatic repulsion between ASP865 and the 1′-cyano group of N-Rem as well as the steric clash between SER861 and the 1′-cyano group in a position where N-Rem reaches after three nucleotide incorporations. They also found that N-Rem at this site greatly weakened the hydrogen bonds of base pairs with its template uracil due to the electrostatic attraction between LYS593 and the 1′-cyano group. Their simulation study showed that the 1′-cyano group on the ribose was essential for remdesivir to inhibit the RdRp function.
Using MD simulations and quantum mechanics/molecular mechanics simulations for SARS-CoV-2 RdRp with an RNA duplex, Aranda et al. reported the detailed mechanisms of the binding and incorporation of natural nucleotides and RemTP [37]. They found that RemTP was preferentially bound to RdRp over ATP, while it was incorporated into the nascent RNA strand with an efficiency only slightly lower than ATP. In addition, they reported that, unlike the simulation results obtained by Zhang et al. [36], no steric clash was detected between N-Rem and the residues of RdRp when the nascent RNA strand was translocated along the exit channel. Instead, they found that N-Rem was trapped at a position where the three nucleotides were incorporated after RemTP. Therefore, they suggested that either non-covalent or transient-covalent bonds between the 1′-cyano group of N-Rem at this position and hydroxyl group of SER861 could act as a trap for the nascent RNA strand and stall the translocation of the duplex.
Luo et al. performed MD simulations to elucidate the nascent-RNA-synthesis inhibition mechanism by remdesivir embedded in the template strand (T-Rem) [38]. Experimental observations have shown that T-Rem inhibits the synthesis of the nascent RNA strand [39]. They revealed that when T-Rem was at the binding site, the translocation of T-Rem was hampered by the hydrogen-bond formation between the 1′-cyano group of T-Rem and the backbone of GLY683 and the steric clash between the 1′-cyano group and the backbone of SER682.
4.2. SARS-CoV-2 RdRp with Other Nucleotide Analogs
Many simulation studies have been conducted on the inhibition mechanism of the RdRp function by nucleotide analogs other than remdesivir. Yuan et al. investigated the inhibitory effect of nucleotide analogs with various 2′ modifications against SARS-CoV-2 RdRp using MD simulations and FEP methods [40]. The nucleotide analogs included 2′-O-methyl uridine triphosphate (OMU-TP), sofosbuvir triphosphate (SFU-TP), 2′-C-methyl cytidine triphosphate (CMC-TP), Gemcitabine triphosphate (GMC-TP), and ara-uridine triphosphate (ARU-TP). Previous experimental studies reported that three of these, OMU-TP, SFU-TP, and CMC-TP, act as effective inhibitors, while GMC-TP and ARU-TP have no inhibitory effects [41][42][43]. They revealed that OMU decreased the binding probability of the subsequent NTP and consequently caused partial chain terminations due to the steric hindrance by its 2′-O-methyl modification. In addition, it was found that the bulky 2′-methyl substitutions in SFU and CMC largely disrupted the binding site, leading to the immediate chain termination. In contrast, GMC and ARU, which have smaller 2′ substitutions such as the fluorine atoms and ara-hydroxyl group, showed marginal effects on the polymerization process upon the incorporation. Their simulation results were consistent with previous experimental results [41][42][43] and elucidated the detailed inhibition mechanisms of 2′ substituted nucleotide analogs against SARS-CoV-2 RdRp.
Li et al. systematically investigated the inhibitory effects of ATP analogs possessing 2′ or 3′ ribose modifications against SARS-CoV-2 RdRp using MD simulations and FEP methods [44]. The analogs included clofarabine triphosphate, didanosine triphosphate, fludarabine triphosphate, vidarabine triphosphate, 2′-amino-2′-deoxyadenosine triphosphate, 2′,3′-didehydro-2′,3′-dideoxyadenosine triphosphate, and cordycepin triphosphate. They found that clofarabine and fludarabine could not form stable binding at the binding site and only had a minor effect on the next nucleotide incorporation into the nascent strand. It was also clarified that vidarabine and 2′-amino-2′-deoxyadenosine could not efficiently inhibit the incorporation of the next substrate, although they could be incorporated into the nascent strand as the substrate. Didanosine, 2′,3′-didehydro-2′,3′-dideoxyadenosine, and cordycepin could also be incorporated into the nascent strand and had the capability to terminate the next nucleotide addition while 2′,3′-didehydro-2′,3′-dideoxyadenosine triphosphate was less competitive than the other two analogs. Therefore, they concluded that substituting the 3′-hydroxyl group with one hydrogen atom would inherently inhibit the next nucleotide addition when it appears at the 3′ terminal of the nascent strand. They proposed that cordycepin and didanosine were promising nucleotide analogs as immediate terminators.
4.3. Overview and Perspective of Molecular Simulations on SARS-CoV-2 RdRp with Nucleotide Analogs
Nucleotide analogs inhibit the RNA replications by interfering with the addition of the next nucleotide (immediate chain termination) [41] or by interfering with the translocation of the nascent RNA strand after the incorporation of three nucleotides (delayed chain termination) [34]. It was shown that nucleotide analogs embedded in the template strand also inhibit the RNA replications (template-dependent inhibition) [39]. The simulation studies presented here elucidated the atomic level mechanisms of the delayed chain termination of remdesivir, immediate chain termination of 2′ and 3′ modified nucleotide analogs, and template-dependent inhibition of remdesivir. Furthermore, these simulation studies also suggested more promising nucleotide analogs for inhibiting the function of RdRp than remdesivir [40][44]. All simulation studies focused on the situation after NTPs or nucleotide analogs are incorporated into the binding site of RdRp. On the other hand, the simulation study described in Section 3 [20] focused on the process by which ligands far from RdRp were incorporated into the binding site and revealed the bucket-brigade transport mechanism of NTPs and nucleotide analogs by lysine residues of RdRp. Overall, the simulation studies presented here help uresearchers in enhancing the understanding on how nucleotide analogs are recognized by RdRp and inhibit the RNA replication at the atomic level.
References
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536-544, 10.1038/s41564-020-0695-z.
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al.Niu, P.Zhan, F.Ma, X.Wang, D.Xu, W.Wu, G.Gao, G. F.Tan, W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727-733, 10.1056/NEJMoa2001017.
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J. W.; Kim, V. N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914-921.e10, https://doi.org/10.1016/j.cell.2020.04.011.
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al.Yuan, M.-L.Zhang, Y.-L.Dai, F.-H.Liu, Y.Wang, Q.-M.Zheng, J.-J.Xu, L.Holmes, E. C.Zhang, Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265-269, https://doi.org/10.1038/s41586-020-2008-3.
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al.Bi, Y.Ma, X.Zhan, F.Wang, L.Hu, T.Zhou, H.Hu, Z.Zhou, W.Zhao, L.Chen, J.Meng, Y.Wang, J.Lin, Y.Yuan, J.Xie, Z.Ma, J.Liu, W. J.Wang, D.Xu, W.Holmes, E. C.Gao, G. F.Wu, G.Chen, W.Shi, W.Tan, W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020, 395, 565-574, https://doi.org/10.1016/S0140-6736(20)30251-8.
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al.Zheng, M.Chen, L.Li, H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766-788, 10.1016/j.apsb.2020.02.008.
- Sanders, J. M.; Monogue, M. L.; Jodlowski, T. Z.; Cutrell, J. B. Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 323, 1824-1836, 10.1001/jama.2020.6019.
- Wang, Y.; Anirudhan, V.; Du, R.; Cui, Q.; Rong, L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2021, 93, 300-310, https://doi.org/10.1002/jmv.26264.
- Venkataraman, S.; Prasad, B. V. L. S.; Selvarajan, R. RNA Dependent RNA Polymerases: Insights from Structure, Function and Evolution. Viruses 2018, 10, 76, https://doi.org/10.3390/v10020076.
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, Y.; Sun, Q.; Ming, Z.; Zhang, L.; et al.Ge, J.Zheng, L.Zhang, Y.Wang, H.Zhu, Y.Zhu, C.Hu, T.Hua, T.Zhang, B.Yang, X.Li, J.Yang, H.Liu, Z.Xu, W.Guddat, L. W.Wang, Q.Lou, Z.Rao, Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779-782, 10.1126/science.abb7498.
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al.Chang, S.Xie, Y.-C.Tian, G.Jiang, H.-W.Tao, S.-C.Shen, J.Jiang, Y.Jiang, H.Xu, Y.Zhang, S.Zhang, Y.Xu, H. E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499-1504, 10.1126/science.abc1560.
- Kirchdoerfer, R. N.; Ward, A. B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 2342, https://doi.org/10.1038/s41467-019-10280-3.
- Subissi, L.; Posthuma, C. C.; Collet, A.; Zevenhoven-Dobbe, J. C.; Gorbalenya, A. E.; Decroly, E.; Snijder, E. J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900-E3909, 10.1073/pnas.1323705111.
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267, https://doi.org/10.3390/cells9051267.
- Elfiky, A. A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2021, 39, 3204-3212, 10.1080/07391102.2020.1761882.
- Zhang, W.-F.; Stephen, P.; Thériault, J.-F.; Wang, R.; Lin, S.-X. Novel Coronavirus Polymerase and Nucleotidyl-Transferase Structures: Potential to Target New Outbreaks. J. Phys. Chem. Lett. 2020, 11, 4430-4435, https://doi.org/10.1021/acs.jpclett.0c00571.
- Itoh, S. G.; Tanimoto, S.; Okumura, H. Dynamic properties of SARS-CoV and SARS-CoV-2 RNA-dependent RNA polymerases studied by molecular dynamics simulations. Chem. Phys. Lett. 2021, 778, 138819, https://doi.org/10.1016/j.cplett.2021.138819.
- Peng, Q.; Peng, R.; Yuan, B.; Zhao, J.; Wang, M.; Wang, X.; Wang, Q.; Sun, Y.; Fan, Z.; Qi, J.; et al.Gao, G. F.Shi, Y. Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2. Cell Rep. 2020, 31, 107774, https://doi.org/10.1016/j.celrep.2020.107774.
- Tanimoto S.; Itoh S. G.; Okumura H. State of the art molecular dynamics simulation studies of RNA-dependent RNA polymerase of SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 10358, 10.3390/ijms231810358.
- Tanimoto, S.; Itoh, S. G.; Okumura, H. “Bucket brigade” using lysine residues in RNA-dependent RNA polymerase of SARS-CoV-2. Biophys. J. 2021, 120, 3615-3627, https://doi.org/10.1016/j.bpj.2021.07.026.
- Warren, T. K.; Jordan, R.; Lo, M. K.; Ray, A. S.; Mackman, R. L.; Soloveva, V.; Siegel, D.; Perron, M.; Bannister, R.; Hui, H. C.; et al.Larson, N.Strickley, R.Wells, J.Stuthman, K. S.Van Tongeren, S. A.Garza, N. L.Donnelly, G.Shurtleff, A. C.Retterer, C. J.Gharaibeh, D.Zamani, R.Kenny, T.Eaton, B. P.Grimes, E.Welch, L. S.Gomba, L.Wilhelmsen, C. L.Donald, N. K.Jonathan, N. E.Elyse, N. R.Jeffrey, K. R.Palacios, G.Doerffler, E.Neville, S.Carra, E.Clarke, M. O.Zhang, L.Lew, W.Ross, B.Wang, W.Chun, K.Wolfe, L.Babusis, D.Park, Y.Stray, K. M.Trancheva, I.Feng, J. Y.Barauskas, O.Xu, Y.Wong, P.Braun, M. R.Flint, M.McMullan, L. K.Chen, S.-S.Fearns, R.Swaminathan, S.Mayers, D. L.Spiropoulou, C. F.Lee, W. A.Nichol, S. T.Cihlar, T.Bavari, S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature 2016, 531, 381-385, https://doi.org/10.1038/nature17180.
- Furuta, Y.; Takahashi, K.; Kuno-Maekawa, M.; Sangawa, H.; Uehara, S.; Kozaki, K.; Nomura, N.; Egawa, H.; Shiraki, K. Mechanism of Action of T-705 against Influenza Virus. Antimicrob. Agents Chemother. 2005, 49, 981-986, https://doi.org/10.1128/AAC.49.3.981-986.2005.
- Peng, Q.; Peng, R.; Yuan, B.; Wang, M.; Zhao, J.; Fu, L.; Qi, J.; Shi, Y. Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir. Innovation 2021, 2, 100080, https://doi.org/10.1016/j.xinn.2021.100080.
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al.Fang, X.Yang, X.Huang, Y.Gao, H.Liu, F.Ge, J.Sun, Q.Yang, X.Xu, W.Liu, Z.Yang, H.Lou, Z.Jiang, B.Guddat, L. W.Gong, P.Rao, Z. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417-428, https://doi.org/10.1016/j.cell.2020.05.034.
- Kokic, G.; Hillen, H. S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P.; et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279, https://doi.org/10.1038/s41467-020-20542-0.
- Naydenova, K.; Muir, K. W.; Wu, L.-F.; Zhang, Z.; Coscia, F.; Peet, M. J.; Castro-Hartmann, P.; Qian, P.; Sader, K.; Dent, K.; et al.Kimanius, D.Sutherland, J. D.Löwe, J.Barford, D.Russo, C. J. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. USA 2021, 118, e2021946118, https://doi.org/10.1073/pnas.2021946118.
- Narayanan, N.; Nair, D. T. Vitamin B12 may inhibit RNA-dependent-RNA polymerase activity of nsp12 from the SARS-CoV-2 virus. IUBMB Life 2020, 72, 2112-2120, https://doi.org/10.1002/iub.2359.
- Ebrahimi, K. S.; Ansari, M.; Moghaddam, M. S. H.; Ebrahimi, Z.; Salehi, Z.; Shahlaei, M.; Moradi, S. In silico investigation on the inhibitory effect of fungal secondary metabolites on RNA dependent RNA polymerase of SARS-CoV-II: A docking and molecular dynamic simulation study. Comput. Biol. Med. 2021, 135, 104613, 10.1016/j.compbiomed.2021.104613.
- Kumar, S.; Sharma, P. P.; Upadhyay, C.; Kempaiah, P.; Rathi, B.; Poonam Multi-targeting approach for nsp3, nsp9, nsp12 and nsp15 proteins of SARS-CoV-2 by Diosmin as illustrated by molecular docking and molecular dynamics simulation methodologies. Methods 2021, 195, 44-56, https://doi.org/10.1016/j.ymeth.2021.02.017.
- Begum, F.; Srivastava, A. K.; Ray, U. Repurposing nonnucleoside antivirals against SARS-CoV2 NSP12 (RNA dependent RNA polymerase): In silico-molecular insight. Biochem. Biophys. Res. Commun. 2021, 571, 26-31, https://doi.org/10.1016/j.bbrc.2021.07.050.
- Ghosh, D.; Ghosh Dastidar, D.; Roy, K.; Ghosh, A.; Mukhopadhyay, D.; Sikdar, N.; Biswas, N. K.; Chakrabarti, G.; Das, A. Computational prediction of the molecular mechanism of statin group of drugs against SARS-CoV-2 pathogenesis. Sci. Rep. 2022, 12, 6241, https://doi.org/10.1038/s41598-022-09845-y.
- Samy, M. N.; Gomaa, A. A.-R.; Attia, E. Z.; Ibrahim, M. A. A.; Desoukey, S. Y.; Kamel, M. S. Flavonoids of Zinnia elegans: Chemical profile and in vitro antioxidant and in silico anti-COVID-19 activities. S. Afr. J. Bot. 2022, 147, 576-585, https://doi.org/10.1016/j.sajb.2022.02.024.
- Lehmann, K. C.; Gulyaeva, A.; Zevenhoven-Dobbe, J. C.; Janssen, G. M. C.; Ruben, M.; Overkleeft, H. S.; van Veelen, P. A.; Samborskiy, D. V.; Kravchenko, A. A.; Leontovich, A. M.; et al.Sidorov, I. A.Snijder, E. J.Posthuma, C. C.Gorbalenya, A. E. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015, 43, 8416-8434, https://doi.org/10.1093/nar/gkv838.
- Gordon, C. J.; Tchesnokov, E. P.; Woolner, E.; Perry, J. K.; Feng, J. Y.; Porter, D. P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785-6797, https://doi.org/10.1074/jbc.RA120.013679.
- Zhang, L.; Zhou, R. Structural Basis of the Potential Binding Mechanism of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase. J. Phys. Chem. B 2020, 124, 6955-6962, https://doi.org/10.1021/acs.jpcb.0c04198.
- Zhang, L.; Zhang, D.; Wang, X.; Yuan, C.; Li, Y.; Jia, X.; Gao, X.; Yen, H.-L.; Cheung, P. P.-H.; Huang, X.; et al. 1′-Ribose cyano substitution allows Remdesivir to effectively inhibit nucleotide addition and proofreading during SARS-CoV-2 viral RNA replication. Phys. Chem. Chem. Phys. 2021, 23, 5852-5863, https://doi.org/10.1039/D0CP05948J.
- Aranda, J.; Wieczór, M.; Terrazas, M.; Brun-Heath, I.; Orozco, M. Mechanism of reaction of RNA-dependent RNA polymerase from SARS-CoV-2. Chem Catal. 2022, 2, 1084-1099, https://doi.org/10.1016/j.checat.2022.03.019.
- Luo, X.; Xu, T.; Gao, X.; Zhang, L. Alternative role of motif B in template dependent polymerase inhibition. Chin. J. Chem. Phys. 2022, 35, 407-412, https://doi.org/10.1063/1674-0068/cjcp2203053.
- Tchesnokov, E. P.; Gordon, C. J.; Woolner, E.; Kocinkova, D.; Perry, J. K.; Feng, J. Y.; Porter, D. P.; Götte, M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020, 295, 16156-16165, https://doi.org/10.1074/jbc.AC120.015720.
- Yuan, C.; Goonetilleke, E. C.; Unarta, I. C.; Huang, X. Incorporation efficiency and inhibition mechanism of 2′-substituted nucleotide analogs against SARS-CoV-2 RNA-dependent RNA polymerase. Phys. Chem. Chem. Phys. 2021, 23, 20117-20128, https://doi.org/10.1039/D1CP03049C.
- Jockusch, S.; Tao, C.; Li, X.; Anderson, T. K.; Chien, M.; Kumar, S.; Russo, J. J.; Kirchdoerfer, R. N.; Ju, J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir. Res. 2020, 180, 104857, https://doi.org/10.1016/j.antiviral.2020.104857.
- Chien, M.; Anderson, T. K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J. J.; Kirchdoerfer, R. N.; Ju, J. Nucleotide Analogues as Inhibitors of SARS-CoV-2 Polymerase, a Key Drug Target for COVID-19. J. Proteome Res. 2020, 19, 4690-4697, https://doi.org/10.1021/acs.jproteome.0c00392.
- Lu, G.; Zhang, X.; Zheng, W.; Sun, J.; Hua, L.; Xu, L.; Chu, X.-J.; Ding, S.; Xiong, W. Development of a Simple In Vitro Assay To Identify and Evaluate Nucleotide Analogs against SARS-CoV-2 RNA-Dependent RNA Polymerase. Antimicrob. Agents Chemother. 2021, 65, e01508-e01520, https://doi.org/10.1128/AAC.01508-20.
- Li, Y.; Zhang, D.; Gao, X.; Wang, X.; Zhang, L. 2′- and 3′-Ribose Modifications of Nucleotide Analogues Establish the Structural Basis to Inhibit the Viral Replication of SARS-CoV-2. J. Phys. Chem. Lett. 2022, 13, 4111-4118, https://doi.org/10.1021/acs.jpclett.2c00087.
