Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Cong Gao.
Bioplastics are polymers made from sustainable bio-based feedstocks. The potential of producing bio-based monomers in microbes has been investigated for decades, their economic feasibility is still unsatisfactory compared with petroleum-derived methods.
- bioplastics
- monomer
- synthetic biology
1. Introduction
Current plastics are mainly synthesized or semi-synthesized from fossil fuels [1]. During this process, greenhouse gas emissions are associated with every stage of the production, application, recycling, and incineration of plastic [2]. As a result, plastic pollution has become one of the most important environmental problems. To eliminate the dependence on fossil fuels and reduce environmental pollution, bioplastics, a substitute for fossil-based plastics, have attracted increasing attention [3,4,5,6][3][4][5][6]. Bioplastics are bio-based and/or biodegradable polymers made from renewable resources, such as cellulose, starch, lignin, etc. [7,8,9][7][8][9]. Compared with fossil-based plastics, bio-based plastics have a lower carbon footprint and exhibit favorable material properties, playing a key role in creating a low-carbon circular economy [10]. In addition, bioplastics can be recycled into raw materials, which improves the efficiency of resource recycling and makes a crucial contribution to resource conservation and environmental protection [11].
According to the annual report (2021) from European Bioplastics, the representative bioplastics included polybutylene adipate terephthalate (PBAT), polylactic acid (PLA), polybutylene succinic acid (PBS), Polyamide (PA), polytrimethylene terephthalate (PTT), and polyethylene terephthalate (PET), which account for about 65% of the total bioplastics market share. Among them, the common monomers included 1,4-butanediol (1,4-BDO), 1,3 propanediol (1,3-PDO), adipic acid, succinic acid, cadaverine, glutaric acid, lactic acid, and terephthalic acid, etc. (Table 1).
Table 1. Applications and market potential of bioplastics monomers.
| Monomers | Applications | Market | References | |||||
|---|---|---|---|---|---|---|---|---|
| 1,4-butanediol | Medical treatment, Food, Chemical industry, Materials | USD 6.19 billion | [12,13,14] | [12][13][14] | ||||
| 1,3 propanediol | Chemical industry, Materials | USD 776.3 million | [15,16,17] | [15][16][17] | ||||
| Succinic acid | Agriculture, Green solvents, Pharmaceuticals, Biodegradable plastics, Materials |
245,000 tons | [18,19] | [18][19] | ||||
| Adipic acid | Chemical industry, Materials | USD 6 billion | [20,21,22] | [20][21][22] | ||||
| Cadaverine | Nylon, Chelating agents, Materials | 220 million | [23,24, | [ | 25,26, | 23 | 27,28] | ][24][25][26][27][28] |
| Glutaric acid | Fine chemicals, Monomers, Building blocks, Materials | NM | [29,30,31] | [29][30][31] | ||||
| Lactic acid | Food, Pharmaceutical, Chemical, Cosmetic industries | 2 million tons | [32,33,34,35] | [32][33][34][35] |
2. Efficient Utilization of Cheap Substrates
There are abundant renewable resources in nature, such as CO2, lignocellulose, glucose, etc., but the efficient utilization of these substrates is challenging. To solve this issue and improve the synthetic efficiency of engineered strains, three main approaches could be developed, including (i) designing substrate utilization pathways; (ii) enhancing substrate utilization capacity; and (iii) optimizing the substrate transformation process (Figure 21).
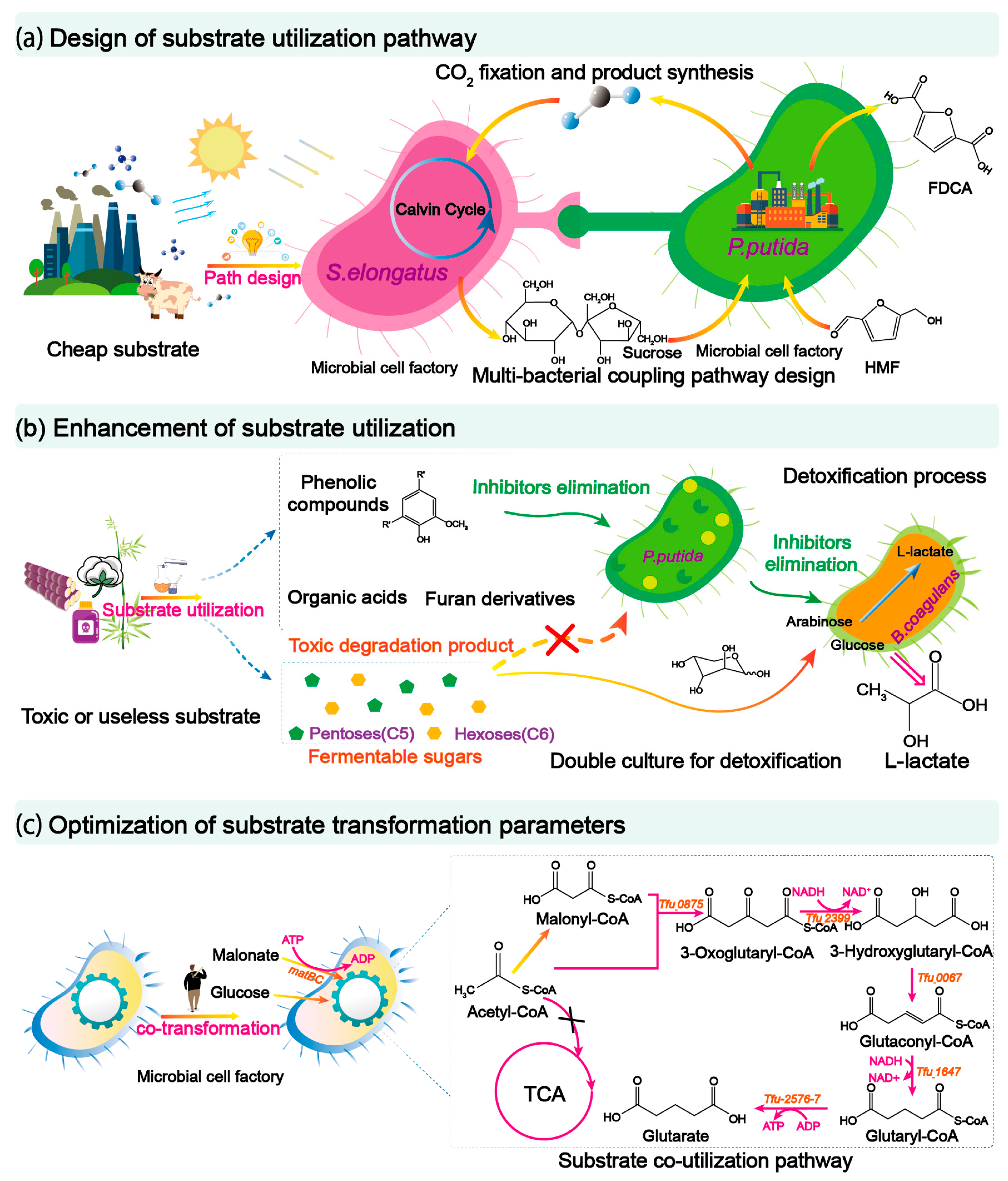
Figure 21. Strategies for efficient utilization of cheap substrates. (a) A multi-bacterial coupling pathway was designed for the utilization of substrates. S. elongatus was used for the fixation of CO2 and the supply of sucrose, while P. putida used sucrose to synthesize 2,5-Furandicarboxylic acid to achieve the utilization of substrates. HMF, 5-hydroxymethylfurfural; FDCA, 2,5-Furandicarboxylic acid. (b) The toxic substrate is utilized through the two-bacterium co-culture system, and P. putida metabolizes the toxic substances in the hydrolysate, promoting the efficient synthesis of L-lactate by B. coagulans, to achieve the detoxification treatment of the toxic substrate and microbial synthesis. (c) By designing the co-utilization of malonate and glucose, the efficient synthesis of glutarate can be realized.
2.1. Designing Substrate Utilization Pathways
According to the types of metabolic pathways, the design of substrate utilization pathways can be divided into five categories: (i) exploiting endogenous substrate utilization pathway; (ii) building artificial substrate utilization pathway; (iii) combining endogenous and artificial substrate utilization pathways; (iv) building orthogonal substrate utilization pathways; (v) coupling multi-bacterial substrate utilization pathways. All these pathway designs can improve the conversion of cheap substrate into high-value-added bio-based monomers.
2.1.1. Exploiting Endogenous Substrate Utilization Pathway
Many bio-based monomers are produced by heterotrophic microorganisms, such as E. coli and yeast, but these microorganisms are difficult to use C1 substrates, such as CO2 and methane. By recruiting microorganisms with native utilization capacity of C1 substrates, the utilization pathway can generate a strong metabolic driving force to maximize the carbon flow towards the desired monomer production. For example, the photoautotrophic microbe S. elongatus could be engineered to produce lysine by promoter characterization and genome-level metabolic pathway engineering. On this basis, different heterogeneous synthetic pathways were introduced in the engineered strain to achieve the direct photosynthetic production of bio-based monomers (such as cadaverine and glutaric acid) from CO2 [113][36]. Similarly, an engineered type II methanotroph, M. trichosporium OB3b, was engineered for the biosynthesis of cadaverine from methane. By introducing lysine decarboxylase, aspartokinase, and meso-diaminopimelate decarboxylase, the final engineered strain could produce 283.63 mg·L−1 of cadaverine directly from methane [49][37].
2.1.2. Building Artificial Substrate Utilization Pathway
For some special substrates, there are no natural substrate utilization pathways for some microorganisms, thus hindering the current progress of developing efficient microbial cell factories to produce desired products. To this end, different artificial pathways can be designed to enable engineered strains to utilize non-metabolic substrates, such as chemical wastes or food wastes, to obtain the desired chemicals. For example, by enzyme mining, multi-enzyme cascade reaction design, and metabolic optimization, a P. taiwanensis VLB120 was engineered to utilize cyclohexane to produce 6-hydroxyhexanoic acid and then further decomposed into adipic acid. After pathway enzyme optimization and bioreactor optimization, the final engineered strain could produce 10.2 g·L−1 adipic acid with cyclohexane as substrate [56][38]. The recycling and reuse of food waste is also a promising alternative to support and promote a circular economy [114][39]. For instance, by modularizing the β-oxidation pathway and ω-oxidation pathway in E. coli, different medium-chain α, ω-dicarboxylic acids could be produced from the waste food oil. The engineered strain could produce 15.26 g·L−1 medium-chain α, ω-dicarboxylic acids using waste food oil as the sole carbon source after adaptive evolution [61][40].
2.1.3. Combining Endogenous and Artificial Substrate Utilization Pathways
Although most of the substrates could be utilized through native or artificial pathways, it is challenging to utilize some substrates by a single microorganism or pathway. Therefore, such substrates can only be converted into desired bio-based monomers by combining natural pathways and artificial pathways. For example, although there are some carboxylases for carbon fixation in E. coli, their efficiencies are quite low [63][41]. To solve this problem, self-assembled cadmium sulfide nanoparticles were developed to improve the CO2 fixation process and a CO2 mitigation switch was designed to strengthen the CO2 mitigation process. By integrating synthetic CO2 fixation and CO2 mitigation modules, the engineered E. coli could produce malate with a yield of 1.48 mol·mol−1 glucose by CO2 sequestration [62][42]. No natural microorganisms can directly use sugar to produce 1,3-PDO. To this end, a de novo 1,3-PDO biosynthesis route from sugar was designed by expanding the homoserine synthesis pathway. Combined with protein engineering and the expression of native alcohol dehydrogenase, the engineered E. coli could directly produce 1,3-PDO from the sugar substrate [64][43].
2.1.4. Building Orthogonal Substrate Utilization Pathways
A common problem during microbial production is that carbon flux leaks into pathways other than the target pathway, which might decrease the yield of desired chemicals. To solve this problem, a strategy termed parallel metabolic pathway engineering was proposed in which one kind of carbon source was designed for desired chemical biosynthesis, while another type of carbon source was engineered for supporting cell growth [72][44]. For example, the metabolic pathways of E. coli were engineered, so that only glucose could be used for cis, cis-muconic acid production, while xylose was designed for restoring cell growth from the Dahms pathway. As a result, the engineered E. coli could produce 4.09 g·L−1 cis, cis-muconic acid with a high yield of 0.31 g·g−1 glucose, by co-utilizing glucose-xylose mixtures [72][44]. Similarly, the production yield of 1,3-PDO from glycerol was always limited by the formation of byproducts. To solve this problem, the endogenous glycerol assimilation pathway was eliminated, and mannitol was fed as a co-substrate to improve glycerol flux towards 1,3-PDO synthesis. As a result, glycerol was redirected mainly for 1,3-PDO biosynthesis and the final engineered strain could obtain a 1,3-PDO yield of 0.76 mol·mol−1 glycerol [65][45].
2.1.5. Coupling Multi-Bacterial Substrate Utilization Pathways
The introduction of a heterogeneous metabolic pathway into a strain often causes cell burden, which affects cell growth performance as well as the synthetic efficiency of material monomers [115][46]. To solve this issue, a co-culture strategy was adopted in which one strain was used for the degradation of complex substrates, while the other strain was designed for the synthesis of monomers. For example, by co-culturing the cellulolytic fungus T. reesei and the lactic acid-producing R. delemar, lactic acid was produced from microcrystalline cellulose using a fungal consortium [74][47]. Synthetic consortia can exhibit advantages compared to pure strain because they can combine the functions of different strains to achieve the direct utilization of some special substrates, such as CO2. For instance, a syntrophic consortium was recently proposed by co-culturing engineered S. elongatus and P. putida. In this system, CO2 can be fixed for producing sucrose by S. elongates, and the produced sucrose could support the growth of P. putida, catalyzing the conversion of 5-Hydroxymethylfurfural to 2,5-Furandicarboxylic acid (Figure 21a) [82][48].
2.2. Enhancing Substrate Utilization Capacity
Designing various metabolic pathways for the utilization of cheap and abundant substrates into high-value-added compounds is necessary. However, not all substrates can be directly utilized without pretreatment [116,117][49][50]. Therefore, it is particularly important to strengthen the utilization capacity of the substrates. To achieve this goal, currently, a total of five main strategies could be summarized: (i) expanding substrate spectrum; (ii) optimizing mass transfer; (iii) depolymerizing substrates; (iv) detoxifying substrates; and (v) enhancing strain growth performance.
2.2.1. Expanding Substrate Spectrum
In addition to traditional five-carbon and six-carbon substrates, some polymeric sugars/substrates are difficult to be utilized by microorganisms [66,118][51][52]. By exploiting technologies for expanding the substrate spectrum in the biotransformation process, the overall production cost could be greatly decreased. For example, the wild-type C. glutamicum strain cannot utilize xylooligosaccharide as the substrate for chemical production. To solve this issue, a cell surface display technique was proposed in which β-xylosidase was expressed on the C. glutamicum cell surface using the PorH anchor protein. Furthermore, the xylose assimilation pathway was also introduced to enable the engineered C. glutamicum to produce 11.6 mM cadaverine using xylooligosaccharide as substrate [50][53]. With the same technology, α-amylase was expressed on the cell surface of E. coli to enable the direct production of 1,2-propanediol (1,2-PDO) and 1,3-PDO from starch [66][51].
2.2.2. Optimizing Mass Transfer
The improvement of microbial substrate utilization ability is often accompanied by the improvement of substrate mass transfer efficiency. To increase the substrate mass transfer efficiency, two main methods could be carried out, including (i) developing a water-organic conversion system to enhance interfacial mass transfer efficiency; and (ii) developing biofilm technology to expand substrate enzyme contact area. For example, paraxylene (PX) and terephthalic acid (TPA) are volatile and almost insoluble in water. To increase the mass transfer efficiency, oleyl alcohol with biocompatibility was recruited as an organic phase for biphasic microbial transformation. As a result, PX can be dissolved in oleyl alcohol and allowed to partition into the aqueous phase with a special partition coefficient. In this biphasic microbial transformation system, 6.9 g·L−1 of TPA could be produced after 46 h bioconversion [83][54]. As extracellular organelles, biofilms can be formed on the cell surface to promote the self-assembly of enzymes or nanoparticles to improve the utilization of substrates [119,120,121,122][55][56][57][58]. To enable the utilization of starch in E. coli, a light-driven CdS-biohybrid system was developed to increase the contact surface area of microbial cell membranes as well as improve the intracellular NADH concentration [123][59].
2.2.3. Depolymerizing Substrates
At present, various strategies for utilizing natural or unnatural substrates have been developed. However, most of these methods are not applicable for substrates with highly polymerized substrates, such as biomass feedstock and polymeric materials. To degrade such substrates, different depolymerization methods, including chemical solvent treatment and physical crushing methods, have been developed [124][60]. For example, an aqueous solvent (NaOH/ChCl:TH/water) was recently designed to efficiently hydrolyze sugarcane bagasse and reduce the degree of polymerization of sugarcane bagasse under mild conditions. After pretreatment, the hydrolysates of sugarcane bagasse could be directly used for microbial adipic acid production with a yield of 0.39 g·g−1 glucose, providing a cost-effective benefit for adipic acid production [57][61]. Similarly, a chemo–microbial hybrid process was carried out in E. coli to convert PET to 2-pyrone-4,6-dicarboxylic acid (PDC) with a yield of up to 96%. Specifically, PET was firstly depolymerized to TPA via microwave-assisted hydrolysis using biomass-derived SiO2 catalysts with thiol functionalization. Then, the produced TPA was used as the substrate for PDC production with a whole-cell conversion strategy [84][62].
2.2.4. Detoxification of Substrates
Substrate detoxification is also helpful to improve the substrate utilization capacity. Lignocellulosic biomass is an attractive and sustainable alternative to petroleum-based feedstock for chemical production. However, it usually needs to be pretreated before they can be efficiently used for the biosynthesis of monomers. During the pretreatment process, a variety of inhibitors were inevitably produced that seriously hinder the growth of microorganisms [125][63]. To solve this problem, a synthetic consortium could be developed to achieve the detoxification treatment of inhibitors. For example, in a synthetic consortium of engineered P. putida and B. coagulans, P. putida was designed to detoxify toxic substrate in lignocellulosic hydrolysate, while B. coagulans was used to capture carbon sources from the hydrolysate to produce 35.8 g·L−1 lactic acid (Figure 21b) [75][64]. A similar study was conducted by building a P. putida and E. coli consisting synthetic consortium. As a result, 1.02 g·L−1 medium-chain polyhydroxyalkanoate was produced using lignocellulosic hydrolysate as the substrate [85][65].
2.2.5. Enhancing Strain Growth Performance
Various factors, such as high concentrations of substrate, substrate toxicity, and environmental stress, can result in insufficient cell growth and decreased production performance. To this end, microbial cell growth could be enhanced by fermentation optimization and cell immobilization. For example, by optimizing the culture conditions, the biomass of C. butyricum L4 could be improved to increase the 1,3-PDO titer to 70.1 g·L−1 [67][66]. Free cells are often affected by complex fermentation environments and cannot be reused, thus increasing production costs. Cell immobilization based on biofilm is a promising strategy to maintain cell activity during fermentation production. For instance, a glycosylated membrane with rhamnose modified surface was constructed to immobilize A. succinogenes for producing succinic acid. With a reduced cell lag stage and high reusability, the biofilm-based cell-immobilized fermentation technology enables the engineered strain with an 18% increased succinic acid titer compared with that of free cell fermentation [86][67]. In addition, the design and modification of the bioreactor to better maintain the stable culture medium resulted in better growth and production of microbial cells, and the concentration of 1,3-PDO was increased to 88.6 g·L−1 [126][68].
2.3. Optimizing Substrate Conversion Process
In addition to the design of substrate utilization pathways and enhancement of substrate utilization capacity, the optimization of the substrate conversion process can also be improved to promote the efficient absorption and utilization of substrates. In general, five strategies could be put forward, such as (i) optimizing substrate transport by overexpressing a specific transporter; (ii) relieving carbon catabolite repression to steadily improve substrate absorption; (iii) co-utilizing multi-substrate to couple cell growth and production; (iv) achieving one-step biotransformation to simplify the substrate utilization process; and (v) strengthening energy supplement to build efficient substrate transport.
2.3.1. Optimizing Substrate Transport
The utilization of substrate can be affected by the efficiency of substrate transport. Therefore, it is particularly important to optimize the substrate transport process. Appropriate overexpression of proteins that transport substrate can enhance the absorption of substrates from extracellular to intracellular. For example, by co-expression of CO2 transport and fixation genes of E. coli using different promoters, the engineered strain could produce 89.4 g·L−1 succinic acid by improving the CO2 fixation process [87][69]. Similarly, by overexpressing glycerol kinase GUT1 in yeast Y. lipolytica, the uptake of glycerol was increased and exhibited a positive effect on cell growth and product synthesis [99][70].
2.3.2. Relieving Carbon Catabolite Repression
Overexpressing substrate transporter can greatly improve the efficiency of the substrate conversion process. However, when multiple substrates, such as glucose and xylose exist together, carbon catabolite repression may occur, resulting in the inhibition of xylose utilization by glucose [127][71]. To solve this issue, three main approaches could be recruited to alleviate the catabolite repression phenomenon. Firstly, deleting carbon repression control (Crc) genes. For instance, the Pseudomonas putida could be engineered to convert ferulate to muconate. However, the existence of glucose, which was used for supplementary energy and cell growth in the medium, always causes carbon catabolite repression, leading to impaired production performance. To this end, the Crc encoding gene was deleted and the resulting strain could produce muconate from ferulate with a doubled yield [128][72]. Secondly, screening xylose catabolic operon XylR mutant. For example, by introducing R121C and P363S into XylR, the xylose catabolism was found to be activated without catabolite repression control. As a result, the engineered E. coli strain exhibited a 50% increase in lactate titer than that of the wild-type strain using a glucose-xylose mixture substrate [76][73]. Thirdly, Knocking out glucose transporter Crr. Due to the metabolite repression of glucose on glycerol, the glucose transporter Crr of K. pneumoniae was knocked out to increase the equivalent of glycerol into the synthesis pathway of 1,3-PDO. The engineered strain exhibited an increased 1,3-PDO titer from 61 to 78 g·L−1, with a glycerol conversion rate reaching 59.5% [68][74].
2.3.3. Co-Utilizing Multi-Substrate
Glucose was mainly used for cell growth and production, while some substrates could strengthen the intracellular reductive force and act as a synthetic precursor. To achieve the co-utilizing of multi-substrate, different substrate utilization pathways can be introduced in one strain to coordinate the co-utilization of substrates. For example, by building glycerol and glucose substrate utilization pathways, the NADH flux and metabolic flux for the production of succinic acid could be improved, as the conversion of glycerol to PEP generates twice as much reducing equivalents mole compared with glucose [88][75]. To obtain more precursors for the synthesis of bio-based monomer, the utilization pathway of malonic acid was introduced into a glutaric acid-producing E. coli. The resulting strain produced more precursor malonyl-CoA compared to the control strain, increasing the glutaric acid titer to 6.3 g·L−1 using both glucose and malonic acid as substrates (Figure 21c) [100][76].
2.3.4. Achieving One-Step Biotransformation
To utilize polymeric substrates, such as starch, cellulose, and xylan, multiple independent biological processes are needed, including the production of hydrolases, hydrolysis of substrates to monosaccharide, and metabolism of monosaccharide to desired products. To decrease overall production costs and simplify the process, one-step biotransformation could be designed to enable the substrate to be saccharified and fermented at the same time [129][77]. For example, by adding glucoamylase into the fermentation system, the liquefied cassava starch was directly used for the production of succinic acid by E. coli NZN111 [130][78]. Commercial enzymes are expensive and their addition for saccharification is not economically viable for the large-scale fermentation process. To solve this problem, some specific strains can be recruited for simultaneous saccharification and fermentation of the substrate due to their hydrolyzing and fermenting capabilities, thereby reducing production costs. For instance, an isolated L. manihotivorans DSM 13343 was used for producing 18.69 g·L−1 lactic acid from a mixed culture with food waste as substrate [77][79].
2.3.5. Strengthening Energy Supplement
In addition, the optimization of energy supply in the biotransformation process enables microorganisms to utilize substrates with reduced energy consumption, increasing the utilization efficiency of substrates. To achieve this goal, the intracellular ATP concentration can be enhanced to compensate for substrate absorption and transport processes that consume energy, such as carbon fixation and xylose transport. For example, by overexpressing the ATP-generating phosphoenolpyruvate carboxykinase combined with the CO2 fixation pathway, the CO2 fixation efficiency for the malic acid biosynthesis pathway was increased by 110% [63][41]. Similarly, the ATP-generating phosphoenolpyruvate carboxykinase could also be co-expressed with the xylose utilization pathway, increasing the titer of succinic acid to 11.13 g·L−1 using corn straw as substrate [89][80].
Through the design of the substrate utilization pathways, the enhancement of utilization capacity, and the optimization of the conversion process, substrate utilization plays an important role in bio-based monomer biosynthesis. The advantages of developing efficient substrate utilization strategies can be summarized in three aspects: (i) Reduce production costs. To improve the market competitiveness of bio-based monomers compared with petrochemical-based monomers, it is very important to develop strategies for efficient utilization of cheap and abundant substrates; (ii) Environmental protection. Domestic garbage often causes resource waste and environmental pollution, and the development of microbial utilization strategies is the key to achieving green biological manufacturing [61][40]; (iii) Microbial synthesis performance improvement. The implementation of a series of strategies, including building orthogonal pathways, relieving carbon catabolite repression, strengthening energy supplements, etc., can enhance the efficiency of engineered strains in bio-based monomer production.
3. Improving Bio-Monomer Synthetic Efficiency
Through the efficient utilization of cheap substrates, the production cost of microbial bio-based materials can be greatly decreased. In addition, to promote the conversion of substrate into target products effectively, the synthetic efficiency of the microbial cell factories can be improved from different regulatory levels, such as enzymes, pathways, and the engineered cell itself. At present, there are three methods to improve the synthetic efficiency of monomers, which include: (i) strengthening key enzyme performance; (ii) optimizing synthetic pathway efficiency; and (iii) regulating cellular metabolic networks (Figure 32).
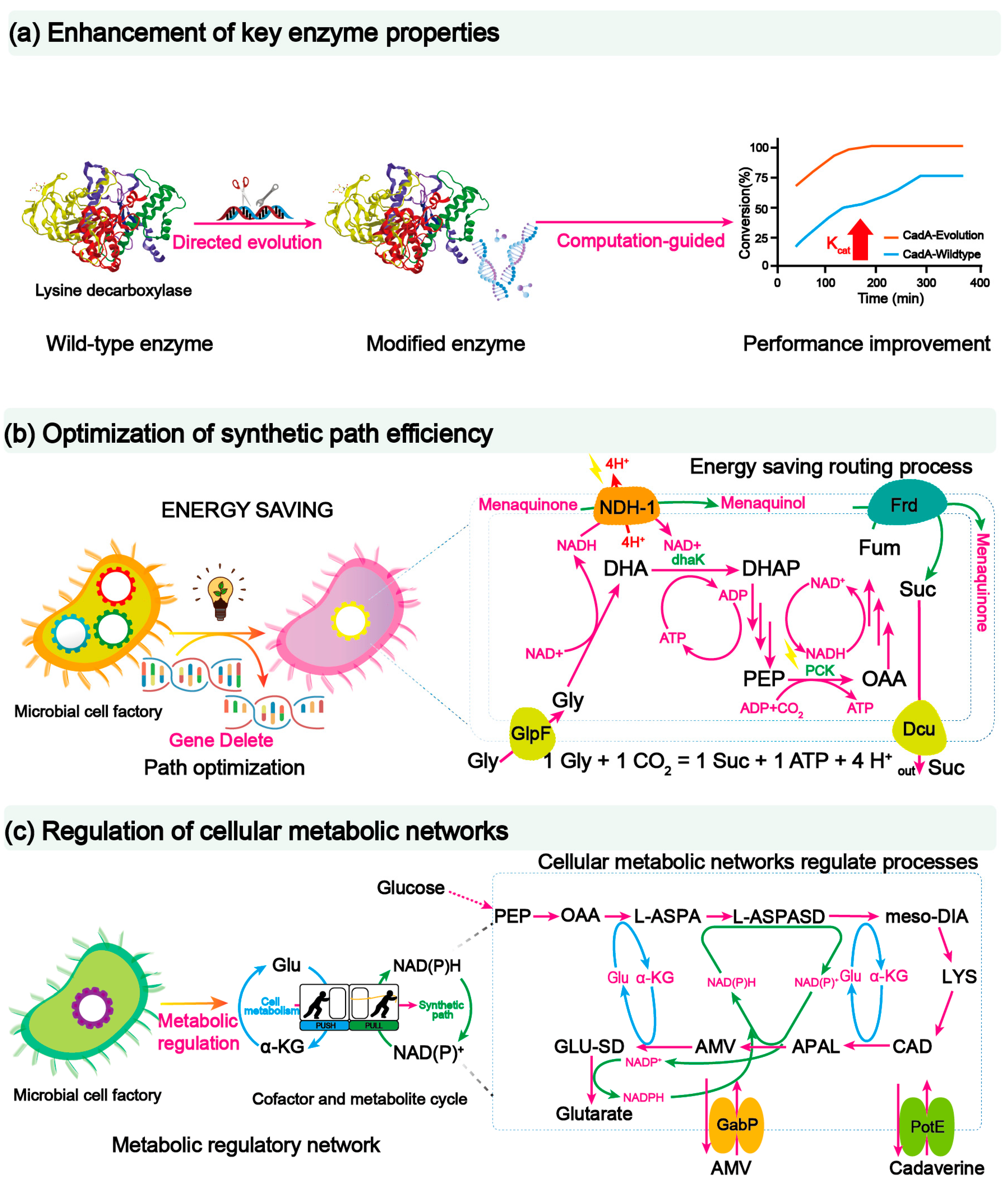
Figure 32. Strategies for improving synthesis efficiency. (a) Aiming at the problem that lysine decarboxylase is not resistant to an alkaline pH environment, directional evolution is introduced to improve the catalytic efficiency of the process and strengthen the performance of L-lysine decarboxylase. (b) The growth of E. coli and the synthesis of succinic acid were improved by constructing a glycerol metabolic pathway with low energy requirements, and the efficiency of the synthetic pathway was increased. Abbreviations: DhaK, ATP-dependent dihydroxyacetone kinase; PCK, phosphoenolpyruvate carboxykinase; Frd, fumarate reductase; Dcu, anaerobic C4-dicarboxylate transporter Dcu; NDH-1, NADH: ubiquinone oxidoreductase I; GlpF, glycerol facilitator; DHA, dihydroxyacetone; DHAP, dihydroxyacetone 3-phosphate; PEP, phosphoenolpyruvate; OAA, oxaloacetate; Fum, fumarate; Suc, succinate; Gly, glycerol. (c) Based on the metabolic drive design, the precise coupling of the metabolic flow between the cell’s central metabolism and the glutarate synthesis pathway was achieved, thus improving the synthesis efficiency of glutarate. Glu, glutamate; α-KG, α-ketoglutarate; PEP, phosphoenolpyruvate; APAL, 5-aminopentanal; AMV, 5-aminovalerate; OAA, oxaloacetate; L-ASPA, L-Aspartate; L-ASPASD, L-Aspartate semialdehyde; meso-DIA, meso-Diaminopimelate; CAD, cadaverine; LYS, lysine; GLU-SD, glutarate semialdehyde.
3.1. Strengthening Key Enzymes Performance
It is a common strategy to improve the production efficiency of monomer microbial cell factories by strengthening the performance of key enzymes. The performance of pathway enzymes could be improved from five aspects: (i) screening heterologous enzymes to obtain more suitable enzymes; (ii) promoting protein correct folding to maximize the enzyme activity; (iii) enhancing protein expression level to drive more metabolic flux; (iv) enzyme directed evolution to modify enzyme properties; and (iii) establishing enzyme recycling technology to reduce the enzymes demand.
3.1.1. Screening Heterologous Enzymes
The performance of pathway enzymes often plays a crucial role in determining the synthetic efficiency and reaction direction in a synthetic pathway. In the microbial synthesis pathway, when the activity of endogenous enzymes is not high enough or there is a lack of enzymes that can play a specific role, it can be optimized by screening for heterologous enzymes with high catalytic efficiency or cofactorless enzymes that can help alleviate the cofactor restriction of endogenous enzymes. For example, malate dehydrogenase (MDH) from different sources was screened and characterized in the engineered M. succiniciproducens. As a result, the MDH from C. glutamicum exhibited the highest enzyme activity and was further used to replace endogenous MDH in the engineered M. succiniciproducens, resulting in succinic acid titer reaching 134.25 g·L−1 [88][75]. In a similar study, the enoate reductase of B. coagulans was introduced into S. cerevisiae. With a three-stage fermentation process optimization, the final engineered strain could directly produce adipic acid using glucose as substrate [58][81].
3.1.2. Promoting Enzyme Folding
The correct folding of enzymes also plays a positive effect on the enhancement of enzyme properties to a certain extent. With correct protein folding, the expression level as well as the catalytic activity of pathway enzymes could be improved to speed up the biosynthesis process. Usually, protein folding can be tuned by optimizing protein expression conditions [103][82], expression host, expression plasmid, signal peptides, molecular tags, and chaperones [131][83]. For example, by introducing zwitterionic peptides into lysine decarboxylase to promote correct folding, the enzymatic activity of engineered lysine decarboxylase was double that of wild-type lysine decarboxylase [54][84]. There are three main elements of the chaperone systems in E. coli, namely, trigger factor, GroEL-GroES, and DnaK-DnaJ-GrpE, which can facilitate the correct folding of polypeptides and prevent the formation of inclusion bodies. By introducing trigger factor chaperone into an engineered E. coli harboring the recombinant 1,2,4-butanetriol pathway, the titer of 1,2,4-butanetriol could be increased from 0.56 g·L−1 to 1.01 g·L−1 [106][85].
3.1.3. Enhancing Protein Expression Level
Enhancing protein expression levels is a simple way to strengthen metabolic efficiency, remove metabolic bottlenecks, and regulate metabolite distribution. Many traditional expression regulatory elements can be used, such as using stronger promoters and high-copy plasmids. For example, to reduce the accumulation and inhibition of cadaverine in engineered E. coli cells, a bi-directional cadaverine transporter PotE was overexpressed using high-copy plasmid to drive more metabolic flux from substrate to cadaverine and the desired product, glutaric acid [101][86]. In enhancing protein expression levels, some species are more suited for genomic modification than others.
3.1.4. Enzymes-Directed Evolution
Pathway enzyme activity could be easily affected by the external as well as the internal environments; thus, it is necessary to modify enzymatic properties, such as catalytic activity, catalytic stability, and stereoselectivity, to improve microbial cell performance. To do so, the simplest and most effective method is to carry out protein-directed evolution [132,133][87][88]. For example, lysine decarboxylase plays a key role in the process of converting lysine decarboxylation to cadaverine, but it is often affected by the alkaline product and the reaction temperature, which hinders its application in industrial production. To this end, the pH stability of lysine decarboxylase was engineered with protein-directed evolution. The resulting mutant M3 exhibited a 6-fold increase in cadaverine production at pH 10.0 than that of the control strain. More importantly, the engineered strain harboring this mutant M3 could produce 418 g·L−1 cadaverine in 15 h, which is the highest titer produced to date [51][89]. Different enzyme properties can be improved simultaneously using protein-directed evolution. For example, both the thermal and alkaline stability of lysine decarboxylase could be improved by combining directed evolution and computation-guided virtual screening. The engineered strain with the best mutant could produce 160.7 g·L−1 cadaverine at 50 °C without pH regulation (Figure 32a) [52][90].
3.1.5. Establishing Enzymes Recycling
Some bio-based monomers, such as glutaric acid and cadaverine, were usually produced by the whole-cell transformation process to obtain high productivity. However, the production cost depends on the input of each batch of enzymes or cells. To solve this problem, various enzyme or cell immobilization strategies could be proposed and exhibited advantages, such as (i) reducing the process cost by recycling cells or enzymes; and (ii) maintaining the catalytic activity of enzymes to resist external environmental stress [102][91]. For example, in a recent study, chitin biopolymer was recruited as a functional material to mediate the immobilization of pyridoxal 5′-phosphate and lysine decarboxylase. Under this design, a continuous biosynthesis of cadaverine without the exogenous addition of pyridoxal 5′-phosphate was achieved [134][92]. A similar study was conducted for glutaric acid biosynthesis. Specifically, a microbial consortium was constructed by immobilizing two types of engineered E. coli cells with colloidal chitin. This efficient microbial platform exhibited good stability and repeatability, resulting in 73.2 g·L−1 glutaric acid accumulation from lysine [102][91].
3.2. Optimizing Synthetic Pathway Efficiency
In addition to the strengthening performance of key enzymes, optimizing the synthetic pathway can also improve the efficiency of monomer biosynthesis to a certain extent. The microbial monomer synthetic pathways are often accompanied by many factors, such as long metabolic reaction steps, by-product accumulation, high energy consumption, etc. Therefore, it is expected that the substrate can be converted into the target products more efficiently by optimizing the synthetic pathway efficiency. At present, five main strategies have been developed, including (i) balancing enzyme expression level; (ii) reconstructing target metabolic flux; (iii) reducing pathway energy consumption; (iv) shortening the spatial distance; and (v) dynamic pathway regulation.
3.2.1. Balancing Enzyme Expression Level
For multiple-enzyme-involved synthetic pathways, metabolic bottlenecks occur frequently because of the differences in catalytic performances and expression levels of pathway enzymes. Such metabolic bottlenecks will lead to the accumulation of intermediate metabolites and further reduce the catalytic efficiency of the metabolic pathway. To this end, the expression level of each enzyme can be regulated to improve the pathway efficiency. Various strategies have been developed to balance enzyme expression levels, such as plasmid optimization [102][91], promoter engineering [135][93], and ribosome binding site engineering [136][94]. For example, a total of seven enzymes involved in the glutaric acid biosynthetic pathway were arranged into plasmids with different copy numbers for combinatorial optimization [102][91]. These pathway enzymes can be further optimized with different strengths of promoters and ribosome binding sites. The final engineered E. coli strain could produce 77.62 g·L−1 glutaric acid with lysine substrate [103][82].
3.2.2. Redirecting Target Metabolic Flux
Microorganisms have evolved extensive regulation and complex interactions between metabolic pathways. As a result, simply introducing a heterologous synthetic pathway for chemical production is often inefficient due to metabolic fugitive effects. Two main approaches can be implemented to redirect the metabolic flux toward target product synthesis. Firstly, increasing precursor concentrations by overexpressing key enzymes in a synthetic pathway. For example, by optimizing the malonyl-coA synthetic pathway to improve the supply of precursors, the malonic acid titer could be increased to 1.62 g·L−1 in S. cerevisiae [107][95]. Similarly, by enhancing the accumulation of 5-aminovaleric acid, the titer of glutaric acid reached 22.7 g·L−1 in the engineered C. glutamicum [40][96]. Secondly, reducing the metabolic loss in the branched pathway by blocking the by-product synthetic pathways. For instance, in a recent study, an autonomous circuit, containing CRISPRi, stationary phase promoter, and protein degradation tag, was successfully constructed for rewiring metabolic flux. By targeting byproduct synthetic pathways, the concentrations of acetic acid, lactic acid, and formic acid in the glutaric acid-producing strain were decreased to 40%, 41%, and 35% compared to that of the control strain, respectively [104][97].
3.2.3. Reducing Pathway Energy Consumption
A third method to improve the synthetic pathway efficiency is to reduce the energy consumption in the target metabolic pathway. By reducing energy consumption, more carbon flux can be rewired to the synthetic pathway in a thermodynamically advantageous manner. To achieve this goal, two main approaches have been developed. Firstly, replacement of high energy consumption pathway. For example, to promote anaerobic succinic acid production, an ATP-dependent dihydroxyacetone kinase from Klebsiella was used to replace the native phosphoenolpyruvate-dependent dihydroxyacetone kinase of E. coli. As a result, the engineered E. coli strain could save one mole of NADH per glycerol to increase succinic acid titer by 282% (Figure 32b) [90][98]. A second way to reduce pathway energy demand is to fine-tune hybrid pathways. By phosphoketolase-mediated non-oxidative glycolysis to accommodate the output NAD(P)H/acetyl-CoA stoichiometries to match the energy demand of the 1,3-butanediol synthesis pathway, the yield of 1,3-butanediol reaches 113% of the theoretical maximum from native metabolism [69][99].
References
- Van Eygen, E.; Feketitsch, J.; Laner, D.; Rechberger, H.; Fellner, J. Comprehensive analysis and quantification of national plastic flows: The case of Austria. Resour. Conserv. Recycl. 2017, 117, 183–194.
- Lee, Y.; Cho, I.J.; Choi, S.Y.; Lee, S.Y. Systems metabolic engineering strategies for non-natural microbial polyester production. Biotechnol. J. 2019, 14, e1800426.
- Bajt, O. From plastics to microplastics and organisms. FEBS Open Bio 2021, 11, 954–966.
- Prata, J.C.; da Costa, J.P.; Lopes, I.; Duarte, A.C.; Rocha-Santos, T. Environmental exposure to microplastics: An overview on possible human health effects. Sci. Total Environ. 2020, 702, 134455.
- Shen, M.C.; Song, B.; Zeng, G.M.; Zhang, Y.X.; Huang, W.; Wen, X.F.; Tang, W.W. Are biodegradable plastics a promising solution to solve the global plastic pollution? Environ. Pollut. 2020, 263, 114469.
- Henderson, L.; Green, C. Making sense of microplastics? Public understandings of plastic pollution. Mar. Pollut. Bull. 2020, 152, 110908.
- Leal, W.; Salvia, A.L.; Bonoli, A.; Saari, U.A.; Voronova, V.; Kloga, M.; Kumbhar, S.S.; Olszewski, K.; De Quevedo, D.M.; Barbir, J. An assessment of attitudes towards plastics and bioplastics in Europe. Sci. Total Environ. 2021, 755, 142732.
- Plastics give and plastics take. Nat. Rev. Mater. 2022, 7, 67. Available online: https://rdcu.be/c4s1i (accessed on 1 December 2022).
- Karan, H.; Funk, C.; Grabert, M.; Oey, M.; Hankamer, B. Green bioplastics as part of a circular bioeconomy. Trends Plant Sci. 2019, 24, 237–249.
- Rosenboom, J.G.; Langer, R.; Traverso, G. Bioplastics for a circular economy. Nat. Rev. Mater. 2022, 7, 117–137.
- Wojnowska-Baryla, I.; Kulikowska, D.; Bernat, K. Effect of bio-based products on waste management. Sustainability 2020, 12, 2088.
- Chocarro-Wrona, C.; de Vicente, J.; Antich, C.; Jimenez, G.; Martinez-Moreno, D.; Carrillo, E.; Montanez, E.; Galvez-Martin, P.; Peran, M.; Lopez-Ruiz, E.; et al. Validation of the 1,4-butanediol thermoplastic polyurethane as a novel material for 3D bioprinting applications. Bioeng. Transl. Med. 2021, 6, e10192.
- Naureen, B.; Haseeb, A.; Basirun, W.J.; Muhamad, F. Recent advances in tissue engineering scaffolds based on polyurethane and modified polyurethane. Mater. Sci. Eng. C-Mater. Biol. Appl. 2021, 118, 111228.
- Li, W.J.; Narancic, T.; Kenny, S.T.; Niehoff, P.J.; O’Connor, K.; Blank, L.M.; Wierckx, N. Unraveling 1,4-butanediol metabolism in Pseudomonas putida KT2440. Front. Microbiol. 2020, 11, 382.
- Yang, M.M.; An, Y.F.; Zabed, H.M.; Guoa, Q.; Yun, J.H.; Zhang, G.Y.; Awad, F.N.; Sun, W.J.; Qi, X.H. Random mutagenesis of Clostridium butyricum strain and optimization of biosynthesis process for enhanced production of 1,3-propanediol. Bioresour. Technol. 2019, 284, 188–196.
- Zabed, H.M.; Zhang, Y.F.; Guo, Q.; Yun, J.H.; Yang, M.M.; Zhang, G.Y.; Qi, X.Y. Co-biosynthesis of 3-hydroxypropionic acid and 1,3-propanediol by a newly isolated Lactobacillus reuteri strain during whole cell biotransformation of glycerol. J. Clean. Prod. 2019, 226, 432–442.
- Zhou, S.F.; Lama, S.; Sankaranarayanan, M.; Park, S. Metabolic engineering of Pseudomonas denitrificans for the 1,3-propanediol production from glycerol. Bioresour. Technol. 2019, 292, 121933.
- Bozell, J.J.; Petersen, G.R. Technology development for the production of biobased products from biorefinery carbohydrates-the US Department of Energy’s “Top 10” revisited. Green Chem. 2010, 12, 539–554.
- Zhou, S.; Zhang, M.; Zhu, L.; Zhao, X.; Chen, J.; Chen, W.; Chang, C. Hydrolysis of lignocellulose to succinic acid: A review of treatment methods and succinic acid applications. Biotechnol. Biofuels Bioprod. 2023, 16, 1.
- Skoog, E.; Shin, J.H.; Saez-Jimenez, V.; Mapelli, V.; Olsson, L. Biobased adipic acid—The challenge of developing the production host. Biotechnol. Adv. 2018, 36, 2248–2263.
- Rios, J.; Lebeau, J.; Yang, T.; Li, S.; Lynch, M.D. A critical review on the progress and challenges to a more sustainable, cost competitive synthesis of adipic acid. Green Chem. 2021, 23, 3172–3190.
- Bart, J.C.J.; Cavallaro, S. Transiting from adipic acid to bioadipic acid. 1, petroleum-based processes. Ind. Eng. Chem. Res. 2015, 54, 1–46.
- Qian, Z.G.; Xia, X.X.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of cadaverine: A five carbon diamine. Biotechnol. Bioeng. 2011, 108, 93–103.
- Melchor-Martinez, E.M.; Castillo, N.T.E.; Macias-Garbett, R.; Lucero-Saucedo, S.L.; Parra-Saldivar, R.; Sosa-Hernandez, J.E. Modern world applications for nano-bio materials: Tissue engineering and COVID-19. Front. Bioeng. Biotechnol. 2021, 9, 597958.
- Wang, J.; Lu, X.L.; Ying, H.X.; Ma, W.C.; Xu, S.; Wang, X.; Chen, K.Q.; Ouyang, P.K. A novel process for cadaverine bio-production using a consortium of two engineered Escherichia coli. Front. Microbiol. 2018, 9, 1312.
- Wang, J.; Mao, J.W.; Tian, W.L.; Wei, G.G.; Xu, S.; Ma, W.C.; Chen, K.Q.; Jiang, M.; Ouyang, P.K. Coproduction of succinic acid and cadaverine using lysine as a neutralizer and CO2 donor with L-lysine decarboxylase overexpressed Escherichia coli AFP111. Green Chem. 2018, 20, 2880–2887.
- Jancewicz, A.L.; Gibbs, N.M.; Masson, P.H. Cadaverine’s functional role in plant development and environmental response. Front. Plant Sci. 2016, 7, 870.
- Ting, W.W.; Huang, C.Y.; Wu, P.Y.; Huang, S.F.; Lin, H.Y.; Li, S.F.; Chang, J.S.; Ng, I.S. Whole-cell biocatalyst for cadaverine production using stable, constitutive and high expression of lysine decarboxylase in recombinant Escherichia coli W3110. Enzym. Microb. Technol. 2021, 148, 109811.
- Rohles, C.M.; Giesselmann, G.; Kohlstedt, M.; Wittmann, C.; Becker, J. Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb. Cell Factories 2016, 15, 154.
- Bhatia, S.K.; Bhatia, R.K.; Yang, Y.H. Biosynthesis of polyesters and polyamide building blocks using microbial fermentation and biotransformation. Rev. Environ. Sci. Bio-Technol. 2016, 15, 639–663.
- Adkins, J.; Jordan, J.; Nielsen, D.R. Engineering Escherichia coli for renewable production of the 5-carbon polyamide building-blocks 5-aminovalerate and glutarate. Biotechnol. Bioeng. 2013, 110, 1726–1734.
- Datta, R.; Henry, M. Lactic acid: Recent advances in products, processes and technologies—A review. J. Chem. Technol. Biotechnol. 2006, 81, 1119–1129.
- Gao, C.; Ma, C.Q.; Xu, P. Biotechnological routes based on lactic acid production from biomass. Biotechnol. Adv. 2011, 29, 930–939.
- Shuklov, I.A.; Dubrovina, N.V.; Kuhlein, K.; Borner, A. Chemo-catalyzed pathways to lactic acid and lactates. Adv. Synth. Catal. 2016, 358, 3910–3931.
- Tian, X.J.; Chen, H.; Liu, H.; Chen, J.H. Recent Advances in lactic acid production by lactic acid bacteria. Appl. Biochem. Biotechnol. 2021, 193, 4151–4171.
- Dookeran, Z.A.; Nielsen, D.R. Systematic engineering of Synechococcus elongatus UTEX 2973 for photosynthetic production of l-lysine, cadaverine, and glutarate. ACS Synth. Biol. 2021, 10, 3561–3575.
- Nguyen, T.T.; Lee, O.K.; Naizabekov, S.; Lee, E.Y. Bioconversion of methane to cadaverine and lysine using an engineered type II methanotroph, Methylosinus trichosporium OB3b. Green Chem. 2020, 22, 7803–7811.
- Bretschneider, L.; Heuschkel, I.; Buehler, K.; Karande, R.; Buehler, B. Rational orthologous pathway and biochemical process engineering for adipic acid production using Pseudomonas taiwanensis VLB120. Metab. Eng. 2022, 70, 206–217.
- Lad, B.C.; Coleman, S.M.; Alper, H.S. Microbial valorization of underutilized and nonconventional waste streams. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab056.
- Li, Y.; Cheng, Z.; Zhao, C.; Gao, C.; Song, W.; Liu, L.; Chen, X. Reprogramming Escherichia coli metabolism for bioplastics synthesis from waste cooking oil. ACS Synth. Biol. 2021, 10, 1966–1979.
- Hu, G.; Zhou, J.; Chen, X.; Qian, Y.; Gao, C.; Guo, L.; Xu, P.; Chen, W.; Chen, J.; Li, Y.; et al. Engineering synergetic CO2-fixing pathways for malate production. Metab. Eng. 2018, 47, 496–504.
- Hu, G.P.; Li, Z.H.; Ma, D.L.; Ye, C.; Zhang, L.P.; Gao, C.; Liu, L.M.; Chen, X.L. Light-driven CO2 sequestration in Escherichia coli to achieve theoretical yield of chemicals. Nat. Catal. 2021, 4, 395–406.
- Chen, Z.; Geng, F.; Zeng, A.P. Protein design and engineering of a de novo pathway for microbial production of 1,3-propanediol from glucose. Biotechnol. J. 2015, 10, 284–289.
- Fujiwara, R.; Noda, S.; Tanaka, T.; Kondo, A. Metabolic engineering of Escherichia coli for shikimate pathway derivative production from glucose-xylose co-substrate. Nat. Commun. 2020, 11, 279.
- Lee, J.H.; Jung, M.Y.; Oh, M.K. High-yield production of 1,3-propanediol from glycerol by metabolically engineered Klebsiella pneumoniae. Biotechnol. Biofuels 2018, 11, 104.
- Zou, W.; Edros, R.; Al-Rubeai, M. The relationship of metabolic burden to productivity levels in CHO cell lines. Biotechnol. Appl. Biochem. 2018, 65, 173–180.
- Scholz, S.A.; Graves, I.; Minty, J.J.; Lin, X.X.N. Production of cellulosic organic acids via synthetic fungal consortia. Biotechnol. Bioeng. 2018, 115, 1096–1100.
- Lin, T.Y.; Wen, R.C.; Shen, C.R.; Tsai, S.L. Biotransformation of 5-hydroxymethylfurfural to 2,5-furandicarboxylic acid by a syntrophic consortium of engineered Synechococcus elongatus and Pseudomonas putida. Biotechnol. J. 2020, 15, 1900357.
- Olson, D.G.; McBride, J.E.; Shaw, A.J.; Lynd, L.R. Recent progress in consolidated bioprocessing. Curr. Opin. Biotechnol. 2012, 23, 396–405.
- Chandel, A.K.; Garlapati, V.K.; Singh, A.K.; Fernandes Antunes, F.A.; da Silva, S.S. The path forward for lignocellulose biorefineries: Bottlenecks, solutions, and perspective on commercialization. Bioresour. Technol. 2018, 264, 370–381.
- Sato, R.; Tanaka, T.; Ohara, H.; Aso, Y. Engineering Escherichia coli for direct production of 1,2-propanediol and 1,3-propanediol from starch. Curr. Microbiol. 2020, 77, 3704–3710.
- Yazawa, K.; Imai, K.; Tamura, Z. Oligosaccharides and polysaccharides specifically utilizable by bifidobacteria. Chem. Pharm. Bull. 1978, 26, 3306–3311.
- Imao, K.; Konishi, R.; Kishida, M.; Hirata, Y.; Segawa, S.; Adachi, N.; Matsuura, R.; Tsuge, Y.; Matsumoto, T.; Tanaka, T.; et al. 1,5-Diaminopentane production from xylooligosaccharides using metabolically engineered Corynebacterium glutamicum displaying beta-xylosidase on the cell surface. Bioresour. Technol. 2017, 245, 1684–1691.
- Luo, Z.W.; Lee, S.Y. Biotransformation of p-xylene into terephthalic acid by engineered Escherichia coli. Nat. Commun. 2017, 8, 15689.
- Jiang, L.; Song, X.G.; Li, Y.F.; Xu, Q.; Pu, J.H.; Huang, H.; Zhong, C. Programming integrative extracellular and intracellular biocatalysis for rapid, robust, and recyclable synthesis of trehalose. ACS Catal. 2018, 8, 1837–1842.
- Nguyen, P.Q.; Botyanszki, Z.; Tay, P.K.R.; Joshi, N.S. Programmable biofilm-based materials from engineered curli nanofibres. Nat. Commun. 2014, 5, 4945.
- Olmez, T.T.; Kehribar, E.S.; Isilak, M.E.; Lu, T.K.; Seker, U.O.S. Synthetic genetic circuits for self-actuated cellular nanomaterial fabrication devices. ACS Synth. Biol. 2019, 8, 2152–2162.
- Wei, W.; Sun, P.Q.; Li, Z.; Song, K.S.; Su, W.Y.; Wang, B.; Liu, Y.Z.; Zhao, J. A surface-display biohybrid approach to light-driven hydrogen production in air. Sci. Adv. 2018, 4, eaap9253.
- Ding, Q.; Liu, Y.D.; Hu, G.P.; Guo, L.; Gao, C.; Chen, X.L.; Chen, W.; Chen, J.; Liu, L.M. Engineering Escherichia coli biofilm to increase contact surface for shikimate and L-malate production. Bioresour. Bioprocess. 2021, 8, 118.
- Gao, C.; Zhang, A.; Chen, K.Q.; Hao, Z.K.; Tong, J.M.; Ouyang, P.K. Characterization of extracellular chitinase from Chitinibacter sp GC72 and its application in GlcNAc production from crayfish shell enzymatic degradation. Biochem. Eng. J. 2015, 97, 59–64.
- Wu, M.; Di, J.; Gong, L.; He, Y.-C.; Ma, C.; Deng, Y. Enhanced adipic acid production from sugarcane bagasse by a rapid room temperature pretreatment. Chem. Eng. J. 2023, 452, 139320.
- Kang, M.J.; Kim, H.T.; Lee, M.W.; Kim, K.A.; Khang, T.U.; Song, H.M.; Park, S.J.; Joo, J.C.; Cha, H.G. A chemo-microbial hybrid process for the production of 2-pyrone-4,6-dicarboxylic acid as a promising bioplastic monomer from PET waste. Green Chem. 2020, 22, 3461–3469.
- Singh, B.; Verma, A.; Pooja; Mandal, P.K.; Datta, S. A biotechnological approach for degradation of inhibitory compounds present in lignocellulosic biomass hydrolysate liquor using Bordetella sp. BTIITR. Chem. Eng. J. 2017, 328, 519–526.
- Zou, L.H.; Ouyang, S.P.; Hu, Y.L.; Zheng, Z.J.; Ouyang, J. Efficient lactic acid production from dilute acid-pretreated lignocellulosic biomass by a synthetic consortium of engineered Pseudomonas putida and Bacillus coagulans. Biotechnol. Biofuels 2021, 14, 227.
- Qin, R.; Zhu, Y.; Ai, M.; Jia, X. Reconstruction and optimization of a Pseudomonas putida-Escherichia coli microbial consortium for mcl-PHA production from lignocellulosic biomass. Front. Bioeng. Biotechnol. 2022, 10, 1023325.
- Gupta, P.; Kumar, M.; Gupta, R.P.; Puri, S.K.; Ramakumar, S.S.V. Fermentative reforming of crude glycerol to 1,3-propanediol using Clostridium butyricum strain L4. Chemosphere 2022, 292, 133426.
- Gao, H.; Wang, J.; Wu, H.; Xin, F.X.; Zhang, W.M.; Jiang, M.; Fang, Y. Biofilm-integrated glycosylated membrane for biosuccinic acid production. ACS Appl. Bio Mater. 2021, 4, 7517–7523.
- Wang, X.L.; Zhou, J.J.; Liu, S.; Sun, Y.Q.; Xiu, Z.L. In situ carbon dioxide capture to co-produce 1,3-propanediol, biohydrogen and micro-nano calcium carbonate from crude glycerol by Clostridium butyricum. Biotechnol. Biofuels Bioprod. 2022, 15, 91.
- Yu, J.H.; Zhu, L.W.; Xia, S.T.; Li, H.M.; Tang, Y.L.; Liang, X.H.; Chen, T.; Tang, Y.J. Combinatorial optimization of CO2 transport and fixation to improve succinate production by promoter engineering. Biotechnol. Bioeng. 2016, 113, 1531–1541.
- Ong, K.L.; Fickers, P.; Lin, C.S.K. Enhancing succinic acid productivity in the yeast Yarrowia lipolytica with improved glycerol uptake rate. Sci. Total Environ. 2020, 702, 134911.
- Gao, C.; Hou, J.; Xu, P.; Guo, L.; Chen, X.; Hu, G.; Ye, C.; Edwards, H.; Chen, J.; Chen, W.; et al. Programmable biomolecular switches for rewiring flux in Escherichia coli. Nat. Commun. 2019, 10, 3751.
- Johnson, C.W.; Abraham, P.E.; Linger, J.G.; Khanna, P.; Hettich, R.L.; Beckham, G.T. Eliminating a global regulator of carbon catabolite repression enhances the conversion of aromatic lignin monomers to muconate in Pseudomonas putida KT2440. Metab. Eng. Commun. 2017, 5, 19–25.
- Sievert, C.; Nieves, L.M.; Panyon, L.A.; Loeffler, T.; Morris, C.; Cartwright, R.A.; Wang, X. Experimental evolution reveals an effective avenue to release catabolite repression via mutations in XylR. Proc. Natl. Acad. Sci. USA 2017, 114, 7349–7354.
- Lu, X.Y.; Ren, S.L.; Lu, J.Z.; Zong, H.; Song, J.; Zhuge, B. Enhanced 1,3-propanediol production in Klebsiella pneumoniae by a combined strategy of strengthening the TCA cycle and weakening the glucose effect. J. Appl. Microbiol. 2018, 124, 682–690.
- Ahn, J.H.; Seo, H.; Park, W.; Seok, J.; Lee, J.A.; Kim, W.J.; Kim, G.B.; Kim, K.J.; Lee, S.Y. Enhanced succinic acid production by Mannheimia employing optimal malate dehydrogenase. Nat. Commun. 2020, 11, 1970.
- Sui, X.; Zhao, M.; Liu, Y.L.; Wang, J.; Li, G.H.; Zhang, X.J.; Deng, Y. Enhancing glutaric acid production in Escherichia coli by uptake of malonic acid. J. Ind. Microbiol. Biotechnol. 2020, 47, 311–318.
- Gao, C.; Guo, L.; Ding, Q.; Hu, G.; Ye, C.; Liu, J.; Chen, X.; Liu, L. Dynamic consolidated bioprocessing for direct production of xylonate and shikimate from xylan by Escherichia coli. Metab. Eng. 2020, 60, 128–137.
- Chen, C.X.; Ding, S.P.; Wang, D.Z.; Li, Z.M.; Ye, Q. Simultaneous saccharification and fermentation of cassava to succinic acid by Escherichia coli NZN111. Bioresour. Technol. 2014, 163, 100–105.
- Rawoof, S.A.A.; Kumar, P.S.; Devaraj, K.; Devaraj, T.; Subramanian, S. Enhancement of lactic acid production from food waste through simultaneous saccharification and fermentation using selective microbial strains. Biomass Convers. Biorefinery 2022, 12, 5947–5958.
- Liu, R.M.; Liang, L.Y.; Chen, K.Q.; Ma, J.F.; Jiang, M.; Wei, P.; Ouyang, P.K. Fermentation of xylose to succinate by enhancement of ATP supply in metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 94, 959–968.
- Raj, K.; Partow, S.; Correia, K.; Khusnutdinova, A.N.; Yakunin, A.F.; Mahadevan, R. Biocatalytic production of adipic acid from glucose using engineered Saccharomyces cerevisiae. Metab. Eng. Commun. 2018, 6, 28–32.
- Wang, J.; Gao, C.; Chen, X.; Liu, L. Engineering the Cad pathway in Escherichia coli to produce glutarate from L-lysine. Appl. Microbiol. Biotechnol. 2021, 105, 3587–3599.
- Schlieker, C. Prevention and reversion of protein aggregation by molecular chaperones in the E. coli cytosol: Implications for their applicability in biotechnology. J. Biotechnol. 2002, 96, 13–21.
- Du, Y.; Pu, Z.J.; Kang, H.; Mi, J.L.; Liu, S.M.; Qi, H.S.; Zhang, L. Zwitterionic peptides encircling-assisted enhanced catalytic performance of lysine decarboxylase for cadaverine biotransformation and mechanism analyses. Chem. Eng. Sci. 2022, 251, 117447.
- Lu, X.Y.; He, S.Y.; Zong, H.; Song, J.; Chen, W.; Bin, Z.G. Improved 1, 2, 4-butanetriol production from an engineered Escherichia coli by co-expression of different chaperone proteins. World J. Microbiol. Biotechnol. 2016, 32, 149.
- Li, W.; Ma, L.; Shen, X.; Wang, J.; Feng, Q.; Liu, L.; Zheng, G.; Yan, Y.; Sun, X.; Yuan, Q. Targeting metabolic driving and intermediate influx in lysine catabolism for high-level glutarate production. Nat. Commun. 2019, 10, 3337.
- Wang, Y.; Xue, P.; Cao, M.; Yu, T.; Lane, S.T.; Zhao, H. Directed evolution: Methodologies and applications. Chem. Rev. 2021, 121, 12384–12444.
- Porter, J.L.; Rusli, R.A.; Ollis, D.L. Directed evolution of enzymes for industrial biocatalysis. Chembiochem 2016, 17, 197–203.
- Gao, S.; Zhang, A.; Ma, D.; Zhang, K.; Wang, J.; Wang, X.; Chen, K. Enhancing pH stability of lysine decarboxylase via rational engineering and its application in cadaverine industrial production. Biochem. Eng. J. 2022, 186, 108548.
- Xi, Y.; Ye, L.D.; Yu, H.W. Enhanced thermal and alkaline stability of L-lysine decarboxylase CadA by combining directed evolution and computation-guided virtual screening. Bioresour. Bioprocess. 2022, 9, 24.
- Gao, C.; Wang, J.P.; Guo, L.; Hu, G.P.; Liu, J.; Song, W.; Liu, L.M.; Chen, X.L. Immobilization of microbial consortium for glutaric acid production from lysine. Chemcatchem 2021, 13, 5047–5055.
- Wei, G.G.; Chen, Y.; Zhou, N.; Lu, Q.H.; Xu, S.; Zhang, A.L.; Chen, K.Q.; Ouyang, P.K. Chitin biopolymer mediates self-sufficient biocatalyst of pyridoxal 5′-phosphate and L-lysine decarboxylase. Chem. Eng. J. 2022, 427, 132030.
- Liang, G.; Zhou, P.; Lu, J.; Liu, H.; Qi, Y.; Gao, C.; Guo, L.; Hu, G.; Chen, X.; Liu, L. Dynamic regulation of membrane integrity to enhance l-malate stress tolerance in Candida glabrata. Biotechnol. Bioeng. 2021, 118, 4347–4359.
- Gao, C.; Tang, W.; Guo, L.; Hu, G.; Liu, J.; Liu, L.; Chen, X. Improving succinate production by engineering oxygen-dependent dynamic pathway regulation in Escherichia coli. Syst. Microbiol. Biomanufacturing 2022, 2, 331–344.
- Li, S.Y.; Fu, W.X.; Su, R.F.; Zhao, Y.Y.; Deng, Y. Metabolic engineering of the malonyl-CoA pathway to efficiently produce malonate in Saccharomyces cerevisiae. Metab. Eng. 2022, 73, 1–10.
- Prell, C.; Busche, T.; Ruckert, C.; Nolte, L.; Brandenbusch, C.; Wendisch, V.F. Adaptive laboratory evolution accelerated glutarate production by Corynebacterium glutamicum. Microb. Cell Factories 2021, 20, 97.
- Gao, C.; Guo, L.; Hu, G.; Liu, J.; Chen, X.; Xia, X.; Liu, L. Engineering a CRISPRi circuit for autonomous control of metabolic flux in Escherichia coli. ACS Synth. Biol. 2021, 10, 2661–2671.
- Yu, Y.; Zhu, X.N.; Xu, H.T.; Zhang, X.L. Construction of an energy-conserving glycerol utilization pathways for improving anaerobic succinate production in Escherichia coli. Metab. Eng. 2019, 56, 181–189.
- Wang, J.; Zhang, R.H.; Zhang, J.L.; Gong, X.Y.; Jiang, T.; Sun, X.X.; Shen, X.L.; Wang, J.; Yuan, Q.P.; Yan, Y.J. Tunable hybrid carbon metabolism coordination for the carbon-efficient biosynthesis of 1,3-butanediol in Escherichia coli. Green Chem. 2021, 23, 8694–8706.
More
