Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 3 by Camila Xu.
Glioblastoma multiforme is a brain tumor with a very unfavorable prognosis, where the vast majority of patients do not survive a year after diagnosis. Arachidonic acid ARA C20:4n-6 in humans is not synthesized de novo but from linoleic acid C18:2n-6 in the polyunsaturated fatty acid (PUFA) biosynthesis pathway. In addition to the cyclooxygenases (COX) pathway, PUFA can be transformed with lipoxygenases (LOX). These enzymes exhibit dioxygenase activity, catalyzing the insertion of a hydroperoxyl group into a PUFA, most commonly ARA 20:4n-6.
- glioblastoma multiforme
- arachidonic acid
- fatty acid
- PUFA
1. Arachidonic Acid Biosynthesis and Glioblastoma Multiforme
1.1. Arachidonic Acid Biosynthesis
ARA C20:4n-6 in humans is not synthesized de novo but from linoleic acid C18:2n-6 in the polyunsaturated fatty acid (PUFA) biosynthesis pathway (Figure 1) [1]. Linoleic acid C18:2n-6 in its activated form, linoleoyl-CoA C18:2n-6, undergoes desaturation with fatty acid desaturase 2 (FADS2)/Δ6-desaturase (D6D), which is accompanied by the formation of γ-linolenoyl-CoA C18:3n-6. Subsequently, the hydrocarbon chain in this fatty acyl-CoA is elongated with two carbons through the elongation of the long-chain fatty acid family members 5 (ELOVL5), accompanied by the formation of dihomo-γ-linolenoyl-CoA C20:3n-6. At the same time, an alternative pathway for the synthesis of dihomo-γ-linolenoyl-CoA C20:3n-6 from linoleoyl-CoA C18:2n-6 is also possible [2]. Linoleoyl-CoA C18:2n-6 is first elongated with ELOVL5 and then desaturated by FADS2. This means that these two enzymes can catalyze the formation of dihomo-γ-linolenoyl-CoA C20:3n-6 in reverse order. In this alternative pathway of PUFA biosynthesis, FADS2 shows activity not of Δ6-desaturase but of Δ8-desaturase. In the latter reaction, the hydrocarbon chain in dihomo-γ-linolenoyl-CoA C20:3n-6 is desaturated with fatty acid desaturase 1 (FADS1)/Δ5-desaturase (D5D), which is accompanied by the production of arachidonyl-CoA C20:4n-6. In the same way as arachidonyl-CoA C20:4n-6, EPA-CoA C20:5n-3 can also be synthesized from α-linolenoyl-CoA C18:3n-3 [1]. Arachidonyl-CoA C20:4n-6 is an activated form of ARA that participates in metabolic pathways, including lipid synthesis pathways. Once synthesized, arachidonyl-CoA C20:4n-6 is used to make lipids, particularly phospholipids. Incorporated into phospholipids, ARA C20:4n-6 is stored and then released by phospholipases A2 (PLA2) as a free fatty acid [3]. Arachidonyl-CoA C20:4n-6 can also be further elongated via elongation of the long-chain fatty acid family members 2 (ELOVL2) and ELOVL5 in a synthesis pathway similar to the synthesis of DHA C22:6n-3 from EPA C20:5n-3 [1][4][5][6].
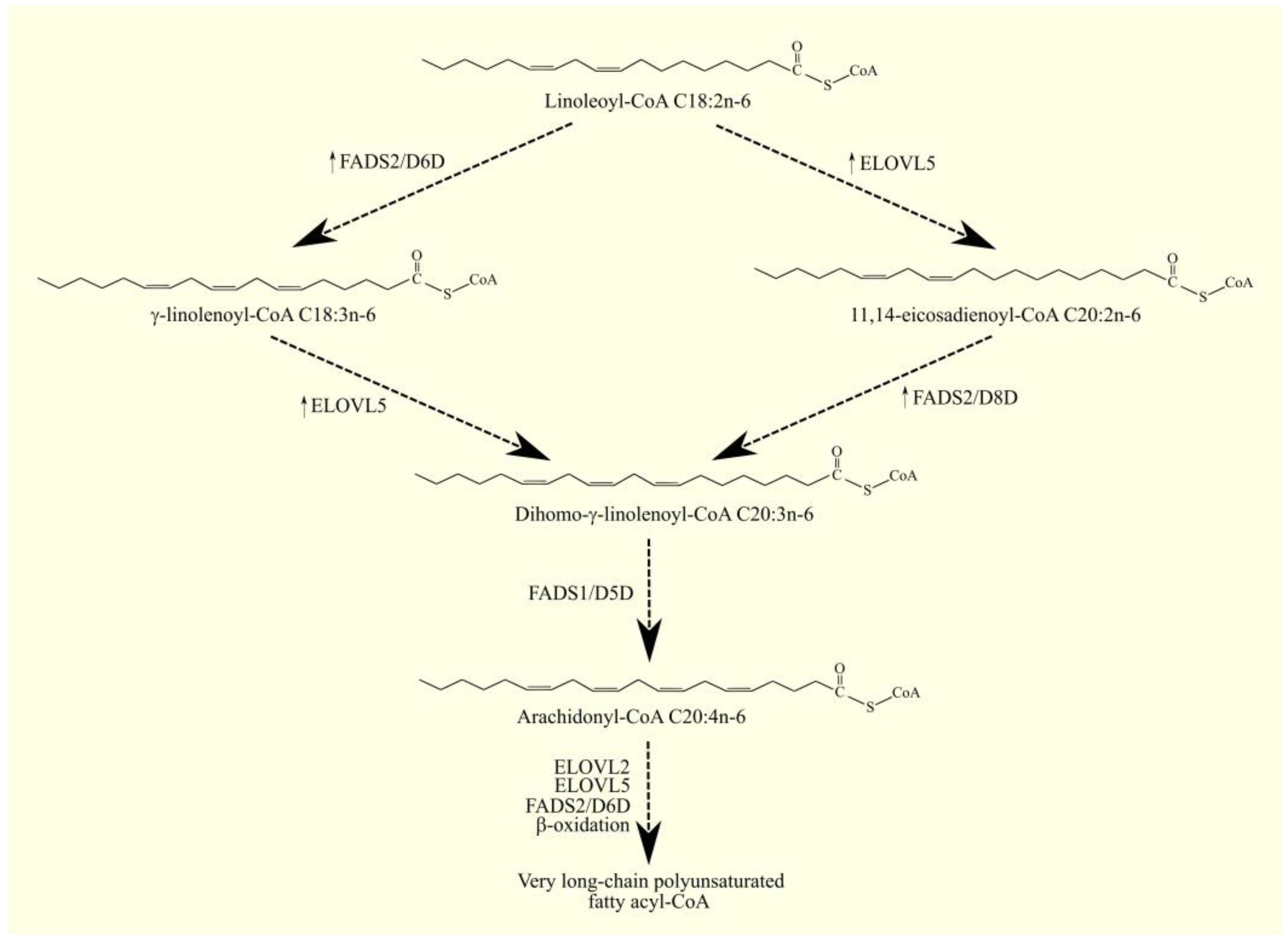
Figure 1. ARA biosynthesis. ARA C20:4n-6 in humans is not synthesized de novo but from linoleic acid C18:2n-6. As linoleoyl-CoA C18:2n-6, this PUFA undergoes desaturation to γ-linolenoyl-CoA C18:3n-6 with FADS2/D6D. This fatty acyl-CoA is then converted to dihomo-γ-linolenoyl-CoA C20:3n-6 with ELOVL5 and, finally, to arachidonyl-CoA C20:4n-6 with FADS1/D5D. Dihomo-γ-linolenoyl-CoA C20:3n-6 can also be formed from linoleoyl-CoA via an alternative pathway. Linoleoyl-CoA C18:2n-6 first undergoes elongation with ELOVL5 and then desaturation with FADS2. The latter enzyme in this pathway exhibits Δ8-desaturase activity. ↑—higher expression of given enzymes in GBM tumor relative to healthy tissue.
1.2. Arachidonic Acid Biosynthesis Pathway in Glioblastoma Multiforme Tumors
Expression of FADS2, an enzyme important for the viability and self-renewal of GBM cancer stem cells [7], is higher in GBM tumors than in healthy brain tissue, according to GEPIA [8] and the transcriptomics analysis performed by Seifert et al. [9]. However, the study showed that FADS2 may have lower expression in tumors than in the peritumoral area in GBM patients [10]. Discrepancies between researchers' results and the data from GEPIA and transcriptomics analysis performed by Seifert et al. may have resulted from studying different groups of patients. FADS2 expression in GBM tumors does not differ between men and women [10]. According to the GEPIA portal, higher FADS2 expression does not affect the prognosis for GBM patients [8]. Studies in GBM models show that FADS2 expression is higher in GBM cancer stem cells than in other GBM cancer cells [7].
The expression of FADS1, which is also important for the viability and self-renewal of GBM cancer stem cells [7], does not differ between GBM tumors and healthy brain tissue, according to GEPIA [8], Seifert et al. [9], and previous results from the research team [10]. According to the GEPIA portal, a higher FADS1 expression does not affect the prognosis for GBM patients [8]. FADS1 expression is higher in GBM cancer stem cells than in other GBM cancer cells [7].
ELOVL5 expression is higher in GBM tumors compared to healthy brain tissue, according to GEPIA [8] and Seifert et al. [9]. However, previous results from the research team did not show significant differences in the expression of ELOVL5 in GBM tumor tissue versus the peritumoral area [11]. Discrepancies between the results and the data from GEPIA and transcriptomics analysis performed by Seifert et al. may have resulted from studying different groups of patients. In addition, it was observed that ELOVL5 expression was lower in GBM tumors in women relative to both the peritumoral area and GBM tumors in men [11]. Higher ELOVL5 expression does not affect the prognosis for GBM patients, according to GEPIA [8]. ELOVL5 expression can be higher in a GBM tumor as a result of hypoxia, as shown by researchers' experiments with U87 MG line cells [11]. This is very important because hypoxia in a GBM tumor also increases the expression of COX-2 [12], an enzyme that converts ARA into prostanoids. This means that hypoxia increases the production of ARA and, at the same time, its conversion into prostanoids.
 Once synthesized, leukotrienes are secreted from the cell. LTC4 is secreted from cells by multidrug resistance-associated proteins (MRP) [57]. In particular, MRP1/ABCC1 [58][59], MRP2/ABCC2 [60][61], MRP3/ABCC3 [62], MRP4/ABCC4 [63], MRP6/ABCC6 [64], MRP7/ABCC10 [65], and MRP8/ABCC11 [66] are responsible for this process. In contrast, OATP1/SLCO1C1 and OATP4 are responsible for the uptake of LTC4, particularly into liver cells where leukotrienes are degraded [67][68]. In contrast, LTB4 transport is still poorly studied; it is known that efflux of LTB4 occurs via MRP4/ABCC4 [63].
Once leukotrienes are secreted outside the cells, they can activate their membrane receptors. LTB4 has two receptors: LTB4R1/BLT1 [46] and LTB4R2/BLT2 [47], the former of which has a 20 times better dissociation constant (Kd) than LTB4R2 in binding LTB4 [47]. With that said, LTB4R2 can be activated by other ARA-derived lipid mediators. These include 12S-HETE, 12R-HETE, 15-HETE, 15-HpETE [69], and 12-HHT [70][71][72]. 12-HHT is formed together with malondialdehyde in a reaction catalyzed by TBXAS1, whose substrate is PGH2 [73][74]. In addition, 12-HHT can be formed independently of TBXAS1 but in smaller amounts [74].
The receptors for cysteinyl-leukotrienes are CysLTR1 [75] and CysLTR2 [76][77]. Both receptors show a 38% similarity in amino acid sequence [76]. CysLTR1 shows a high affinity for LTD4 and low affinity for LTC4 and LTE4, and it shows no affinity at all for LTB4 [75]. CysLTR2 has the best affinity for LTC4 and LTD4 and a very low affinity for LTE4, and it shows no affinity at all for LTB4 [76][77]. A receptor specific for LTE4 is 2-oxoglutarate receptor 1 (OXGR1)/GPR99 [78], which is also the receptor for 2-oxoglutarate. This receptor has a lower affinity for LTC4 and LTD4. Another identified receptor for cysteinyl-leukotrienes specifically for LTC4 and LTD4 is G protein-coupled receptor 17 (GPR17) [79], which is also activated by uridine diphosphate (UDP), UDP-glucose, and UDP-galactose [79]. Further studies have not confirmed that GPR17 is a receptor for UDP, LTC4, and LTD4 [80][81]. This receptor can, independently of its ligand, downregulate CysLTR1 [82], which means it can reduce the action of cysteinyl leukotrienes.
Leukotrienes can be inactivated and excreted. LTB4 is oxidized to 12-oxo-LTB4 with 12-hydroxyeicosanoid dehydrogenase (12-HEDH)/PTGR1 [83][84][85]. This enzyme is also involved in prostaglandin degradation [86]. Subsequently, 12-oxo-LTB4 is reduced with the formation of 12-oxo-10,11-dihydro-LTB4 with an enzyme with Δ10-reductase activity [87]. 12-oxo-10,11-dihydro-LTB4 can then be converted to 10,11-dihydro-LTB4 and 10,11-dihydro-12-epi-LTB4, which undergo ω-oxidation, β-oxidation, or elongation [83]; compounds formed after ω-oxidation and β-oxidation are excreted in the feces [88] and urine [89] as ω-carboxymetabolites of LTB4. HETE are similarly degraded, such as 12-HETE with the formation of 10,11-dihydro-12-HETE and 10,11-dihydro-12-oxo-ETE [90]. Cysteinyl-leukotrienes are first converted to LTE4 [91]; this leukotriene then undergoes ω-oxidation with the formation of ω-carboxy-tetranor-dihydro-LTE4, which is eliminated in the feces and urine.
Once synthesized, leukotrienes are secreted from the cell. LTC4 is secreted from cells by multidrug resistance-associated proteins (MRP) [57]. In particular, MRP1/ABCC1 [58][59], MRP2/ABCC2 [60][61], MRP3/ABCC3 [62], MRP4/ABCC4 [63], MRP6/ABCC6 [64], MRP7/ABCC10 [65], and MRP8/ABCC11 [66] are responsible for this process. In contrast, OATP1/SLCO1C1 and OATP4 are responsible for the uptake of LTC4, particularly into liver cells where leukotrienes are degraded [67][68]. In contrast, LTB4 transport is still poorly studied; it is known that efflux of LTB4 occurs via MRP4/ABCC4 [63].
Once leukotrienes are secreted outside the cells, they can activate their membrane receptors. LTB4 has two receptors: LTB4R1/BLT1 [46] and LTB4R2/BLT2 [47], the former of which has a 20 times better dissociation constant (Kd) than LTB4R2 in binding LTB4 [47]. With that said, LTB4R2 can be activated by other ARA-derived lipid mediators. These include 12S-HETE, 12R-HETE, 15-HETE, 15-HpETE [69], and 12-HHT [70][71][72]. 12-HHT is formed together with malondialdehyde in a reaction catalyzed by TBXAS1, whose substrate is PGH2 [73][74]. In addition, 12-HHT can be formed independently of TBXAS1 but in smaller amounts [74].
The receptors for cysteinyl-leukotrienes are CysLTR1 [75] and CysLTR2 [76][77]. Both receptors show a 38% similarity in amino acid sequence [76]. CysLTR1 shows a high affinity for LTD4 and low affinity for LTC4 and LTE4, and it shows no affinity at all for LTB4 [75]. CysLTR2 has the best affinity for LTC4 and LTD4 and a very low affinity for LTE4, and it shows no affinity at all for LTB4 [76][77]. A receptor specific for LTE4 is 2-oxoglutarate receptor 1 (OXGR1)/GPR99 [78], which is also the receptor for 2-oxoglutarate. This receptor has a lower affinity for LTC4 and LTD4. Another identified receptor for cysteinyl-leukotrienes specifically for LTC4 and LTD4 is G protein-coupled receptor 17 (GPR17) [79], which is also activated by uridine diphosphate (UDP), UDP-glucose, and UDP-galactose [79]. Further studies have not confirmed that GPR17 is a receptor for UDP, LTC4, and LTD4 [80][81]. This receptor can, independently of its ligand, downregulate CysLTR1 [82], which means it can reduce the action of cysteinyl leukotrienes.
Leukotrienes can be inactivated and excreted. LTB4 is oxidized to 12-oxo-LTB4 with 12-hydroxyeicosanoid dehydrogenase (12-HEDH)/PTGR1 [83][84][85]. This enzyme is also involved in prostaglandin degradation [86]. Subsequently, 12-oxo-LTB4 is reduced with the formation of 12-oxo-10,11-dihydro-LTB4 with an enzyme with Δ10-reductase activity [87]. 12-oxo-10,11-dihydro-LTB4 can then be converted to 10,11-dihydro-LTB4 and 10,11-dihydro-12-epi-LTB4, which undergo ω-oxidation, β-oxidation, or elongation [83]; compounds formed after ω-oxidation and β-oxidation are excreted in the feces [88] and urine [89] as ω-carboxymetabolites of LTB4. HETE are similarly degraded, such as 12-HETE with the formation of 10,11-dihydro-12-HETE and 10,11-dihydro-12-oxo-ETE [90]. Cysteinyl-leukotrienes are first converted to LTE4 [91]; this leukotriene then undergoes ω-oxidation with the formation of ω-carboxy-tetranor-dihydro-LTE4, which is eliminated in the feces and urine.
 15-HpETE is transformed into many lipid mediators. It can be transformed into 15-HETE, which is an activator of PPARγ [97] and G2A/GPR132 [35]. 15-HpETE can be converted to 13R-hydroxy-14S,15S-epoxyeicosa-5Z,8Z,11Z-trienoic acid (14,15-HxB3 13R), 11S-hydroxy-14S,15S-epoxy-5Z,8Z,12E-eicosatrienoic acid (14,15-HxA3 11S), and 15-oxo-ETE [18][119]. 14,15-HxA3 11S, analogous to HxA3, can be conjugated with glutathione. This produces 14,15-HxA3-C 11S and cysteinyl-14,15-HxA3 11S, having conjugated glutathione without further amino acids, which is analogous to that of cysteinyl-leukotriene [119].
15-HpETE can also be converted to eoxins [120], which are isomers of leukotrienes.
15-HpETE can also be converted to lipoxins with 5-LOX [42], resulting in the formation of 5S,15S-dihydroperoxyeicosatetraenoic acid (5,15-diHpETE), and then converted to LXA4 or LXB4 [121]. 5-HpETE can also be converted with 15-LOX-1/ALOX15 into 5,15-diHpETE and, via the same pathway, be converted into LXA4 or LXB4 [121]. 15-HETE can be converted to LXA4 with 5-LOX/ALOX5 [122]. Lipoxins can also be formed from LTA4, which is processed by 15-LOX-1/ALOX15 or 12-LOX [121][123].
LXA4 is a lipid mediator with biological activity whose receptors are lipoxin A4 receptor (ALX)/formyl peptide receptor type 2 (FPR2) [124][125], aryl hydrocarbon receptor (AHR) [126], and estrogen receptors subtypes alpha (ERα) [127], the former of which is not a receptor for LXB4 [124]. The ALX/FPR2 receptor is responsible for the anti-inflammatory properties of lipoxins.
There are also cysteinyl lipoxins, which, just like cysteinyl leukotrienes, are lipoxins with conjugated glutathione at carbon 6 [122]. They are synthesized from 15-HETE, from which, with the participation of 5-LOX/ALOX5, 15-hydroxy-5,6-epoxy-eicosatetraenoic acid is formed, a compound similar in structure to LTA4. The epoxy group from these two compounds is converted to a hydroxyl group and conjugated glutathione [122]. However, it is not known whether cysteinyl lipoxins are essential lipid mediators or merely arise as a result of the nonspecificity of enzymes conjugating glutathione to various compounds.
15-HpETE is transformed into many lipid mediators. It can be transformed into 15-HETE, which is an activator of PPARγ [97] and G2A/GPR132 [35]. 15-HpETE can be converted to 13R-hydroxy-14S,15S-epoxyeicosa-5Z,8Z,11Z-trienoic acid (14,15-HxB3 13R), 11S-hydroxy-14S,15S-epoxy-5Z,8Z,12E-eicosatrienoic acid (14,15-HxA3 11S), and 15-oxo-ETE [18][119]. 14,15-HxA3 11S, analogous to HxA3, can be conjugated with glutathione. This produces 14,15-HxA3-C 11S and cysteinyl-14,15-HxA3 11S, having conjugated glutathione without further amino acids, which is analogous to that of cysteinyl-leukotriene [119].
15-HpETE can also be converted to eoxins [120], which are isomers of leukotrienes.
15-HpETE can also be converted to lipoxins with 5-LOX [42], resulting in the formation of 5S,15S-dihydroperoxyeicosatetraenoic acid (5,15-diHpETE), and then converted to LXA4 or LXB4 [121]. 5-HpETE can also be converted with 15-LOX-1/ALOX15 into 5,15-diHpETE and, via the same pathway, be converted into LXA4 or LXB4 [121]. 15-HETE can be converted to LXA4 with 5-LOX/ALOX5 [122]. Lipoxins can also be formed from LTA4, which is processed by 15-LOX-1/ALOX15 or 12-LOX [121][123].
LXA4 is a lipid mediator with biological activity whose receptors are lipoxin A4 receptor (ALX)/formyl peptide receptor type 2 (FPR2) [124][125], aryl hydrocarbon receptor (AHR) [126], and estrogen receptors subtypes alpha (ERα) [127], the former of which is not a receptor for LXB4 [124]. The ALX/FPR2 receptor is responsible for the anti-inflammatory properties of lipoxins.
There are also cysteinyl lipoxins, which, just like cysteinyl leukotrienes, are lipoxins with conjugated glutathione at carbon 6 [122]. They are synthesized from 15-HETE, from which, with the participation of 5-LOX/ALOX5, 15-hydroxy-5,6-epoxy-eicosatetraenoic acid is formed, a compound similar in structure to LTA4. The epoxy group from these two compounds is converted to a hydroxyl group and conjugated glutathione [122]. However, it is not known whether cysteinyl lipoxins are essential lipid mediators or merely arise as a result of the nonspecificity of enzymes conjugating glutathione to various compounds.
2. Lipoxygenases and Arachidonic Acid in Glioblastoma Multiforme
2.1. Lipoxygenases Pathway
In addition to the COX pathway, PUFA can be transformed with LOX. These enzymes exhibit dioxygenase activity, catalyzing the insertion of a hydroperoxyl group into a PUFA, most commonly ARA 20:4n-6. Hydroperoxyeicosatetraenoic acids (HpETE) are then formed from ARA 20:4n-6, which are further processed in the lipoxygenase pathway. The names of LOX enzymes are related to their sites of formation and the configuration of the hydroperoxyl group in ARA 20:4n-6. In humans, there are six LOX:-
epidermal lipoxygenase 3/arachidonate lipoxygenase 3 (eLOX3/ALOXE3),
-
5-lipoxygenase/arachidonate 5-lipoxygenase (5-LOX/ALOX5),
-
12S-lipoxygenase/arachidonate 12-lipoxygenase, 12S type (12S-LOX/ALOX12),
-
12R-lipoxygenase/arachidonate 12-lipoxygenase, 12R type (12R-LOX/ALOX12B),
-
15-lipoxygenase-1/arachidonate 15-lipoxygenase (15-LOX-1/ALOX15), also known as 12/15-LOX, and
-
15-lipoxygenase-2/arachidonate 15-lipoxygenase type B (15-LOX-2/ALOX15B).
2.1.1. Epidermal Lipoxygenase 3
The ALOXE3 gene forms a gene cluster on 17p13.1 together with other LOX [14]. The highest expression of the ALOXE3 gene is found in the skin [14][17]; very low expression of this gene is found in the brain, placenta, pancreas, ovary, and testis. eLOX3/ALOXE3 shows no significant activity against ARA 20:4n-6 or linoleic acid C18:2n-6 [18], which is related to the low availability of molecular oxygen in the active center of this enzyme [19]. For this reason, the processing of ARA 20:4n-6 by eLOX3/ALOXE3 is very inefficient, but eLOX3/ALOXE3 can exhibit dioxygenase activity to ARA 20:4n-6. eLOX3/ALOXE3 has hydroperoxide isomerase activity [18]. eLOX3/ALOXE3 converts HpETE into hydroxy-epoxyeicosatrienoic acid, which is the main product of eLOX3/ALOXE3 activity. eLOX3/ALOXE3 also converts HpETE into oxo-eicosatetraenoic acid (oxo-ETE)/ketoeicosatetraenoic acid (KETE) [18][20]. 15S-HpETE is converted by eLOX3/ALOXE3 into either 13R-hydroxy-14S,15S-epoxyeicosa-5Z,8Z,11Z-trienoic acid or 15-oxo-ETE [18]. eLOX3/ALOXE3 also converts 12S-HpETE into hepoxilin A3 (HxA3), HxB3 [18][21], or 12-oxo-ETE [22][23]. On the other hand, 12R-HpETE is converted by eLOX3/ALOXE3 into either 11,12-bis-epi-HxA3 or 12-oxo-ETE [18]. In addition, eLOX3/ALOXE3 shows activity to 5-HpETE and other HpETEs [20]. Because HETE and oxo-ETE [24] as well as hepoxilins [25] exhibit biological activity, eLOX3/ALOXE3 affects biological and pathological processes, particularly in the skin, where expression of this enzyme is highest. For this reason, mutations in the ALOXE3 gene lead to ichthyosis [26][27][28].2.1.2. 5-Lipoxygenase
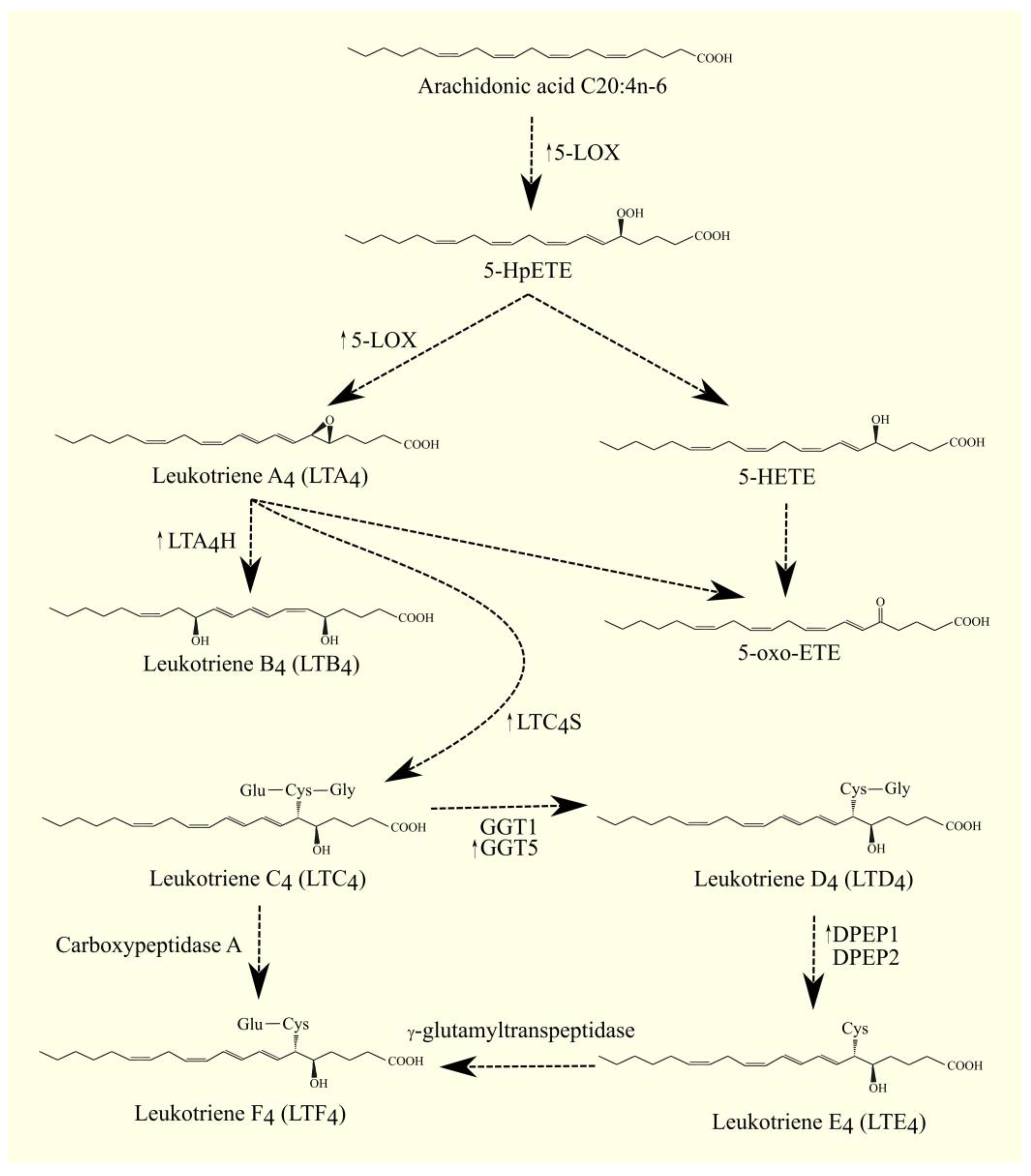
The best-studied LOX is 5-LOX/ALOX5. The highest expression of 5-LOX/ALOX5 is found in the bone marrow, appendix, lung, urinary bladder, spleen, and lymph node [17]. This enzyme converts ARA 20:4n-6 to 5S-hydroperoxyeicosatetraenoic acid (5-HpETE) and then to leukotriene A4 (LTA4) [29]. Importantly, 5-lipoxygenase-activating protein (FLAP)/ALOX5AP is required for the activity of 5-LOX/ALOX5. FLAP/ALOX5AP is a substrate carrier [30][31]. 5-HpETE is an activator of PPARα [32]; for this reason, if it is not converted to other lipid mediators, then it will activate this nuclear receptor. Subsequently, LTA4 is converted to other lipid mediators, in particular to other leukotrienes. LTA4 can also undergo spontaneous conversion to 5,6-diHETE, 5,12-diHETE, and 5-oxo-ETE [33]. In turn, 5-HpETE is converted to 5-hydroxyeicosatetraenoic acid (5-HETE) with glutathione peroxidase [34]. The identified receptor for 5-HETE is G2A/GPR132 [35]; this receptor is also activated by other lipid mediators, such as various HETE and 9-HODE. 5-oxo-ETE can also be formed from 5-HETE with the participation of an enzyme with 5-hydroxyeicosanoid dehydrogenase (5-HEDH) activity [36][37][38]. 5-oxo-ETE is an important lipid mediator with a receptor oxoeicosanoid receptor 1 (OXER1)/GPR99 [39][40][41]. LTA4 is a precursor for the production of other leukotrienes and lipoxins; it is converted to lipoxins in a reaction catalyzed by 12-LOX or 15-LOX [42]. LTA4 can also be converted to LTB4 by LTA4 hydrolase (LTA4H) [43][44]. LTA4H also has aminopeptidase activity unrelated to the production of leukotrienes [44]; this activity is important in moderating the immune response [45]. LTB4 has its own membrane receptors: LTB4R1/BLT1 [46] and LTB4R2/BLT2 [47]. Inside the cell, LTA4 and LTB4 activate PPARα, by which these leukotrienes can exert anti-inflammatory effects [32][48]. Glutathione can be attached to LTA4 by LTC4 synthase (LTC4S) (Figure 2) [49][50]. LTC4 is then formed. LTC4S combines with 5-LOX and FLAP to increase the efficiency of LTC4 production with ARA 20:4n-6 [51]. Subsequently, amino acids from the conjugated glutathione in LTC4 can be removed. As a consequence of this, LTC4 is converted into other leukotrienes, namely LTD4, LTE4, and LTF4. All of these leukotrienes, together with LTC4, form a group called cysteinyl leukotrienes. LTD4 is then formed from LTC4 with the involvement of γ-glutamyltransferase 1 (GGT1) and γ-glutamyltransferase 5 (GGT5) [52]. Subsequently, LTD4 can be converted to LTE4 with the participation of dipeptidase 1 (DPEP1) and dipeptidase 2 (DPEP2) [53][54]. LTC4 can also be converted to LTF4 with the participation of carboxypeptidase A [55]. Amino acids can be attached back to cysteine in cysteinyl leukotriene, as exemplified by the conversion of LTE4 to LTF4 with the participation of an enzyme with γ-glutamyltranspeptidase activity [56]. LTF4, however, has a much weaker effect than LTE4, and the latter reaction can be considered an inactivation of LTE4.
Figure 2. 5-LOX pathway. ARA C20:4n-6 is converted to 5-HpETE with 5-LOX. This enzyme also catalyzes the next step in leukotriene biosynthesis. It converts 5-HpETE into LTA4, which can then be converted into LTB4 with LTA4H, into LTC4 with LTC4S, or into 5-oxo-ETE. 5-HpETE can also be converted to 5-oxo-ETE. LTC4 can be converted to other cysteinyl leukotrienes. LTC4 can be converted to LTF4 with the involvement of carboxypeptidase A or to LTD4 with the involvement of GGT1 and GGT5. Subsequently, LTD4 can be converted into LTE4 with the participation of DPEP1 and DPEP2, and then converted into LTF4 with γ-glutamyltranspeptidase. ↑—higher expression of given enzymes in GBM tumor relative to healthy tissue.
2.1.3. 12S-Lipoxygenase
ALOX12 gene expression is found in the esophagus and skin [17]. 12S-LOX/ALOX12 can participate in the conversion of LTA4 into lipoxins [42], but the best-described activity of 12S-LOX/ALOX12 is to catalyze the insertion of a hydroperoxyl group into ARA 20:4n-6 at position 12—12S-HpETE is then formed [92]—the compound which can also be formed with 15-LOX-1/ALOX15 [93]. 12S-LOX can convert dihomo-γ-linolenic acid to 12S-hydroxy-8Z,10E,14Z-eicosatrienoic acid (12S-HETrE) [94][95]. In contrast, linoleic acid C18:2n-6 is not a substrate for 12S-LOX/ALOX12 [94]. 12S-HpETE can be converted to 12S-HETE, whose receptors are G protein-coupled receptor 31 (GPR31) [96] and G2A/GPR132 [35]. 12S-HETE also activates PPARγ [97], as 12S-HpETE [18] and 12S-HETE can be converted to 12-oxo-ETE [87], a PPARγ ligand and activator [22]. 12-oxo-ETE can be converted back to 12S-HETE with an enzyme with 12-oxo-ETE reductase activity [98]. 12S-HpETE can be converted to HxA3 (8-hydroxy-11,12-epoxyeicosatrienoic acid) or HxB3 (10-hydroxy-11,12-epoxyeicosatrienoic acid) with enzymes with hepoxilin synthase activity, for example, heme, as shown by experiments on hemoglobin and hemin [99][100]. Hepoxilin synthase activity is also demonstrated by eLOX3/ALOXE3, 12S-LOX/ALOX12, and 15-LOX-1/ALOX15, as shown by experiments on human, rat, and mouse models [18][21][101][102]. Then, HxA3 may bind glutathione via glutathione S-transferase at position 11 [103][104]. HxA3 then gives rise to 11-glutathionyl-HxA3, or otherwise HxA3-C. HxB3 is not subject to such modification [105]. HxA3-C can be produced in the brain and may be a neuromodulator [106]. Like cysteinyl-leukotrienes, HxA3-C can be converted to other cysteinyl-hepoxilins [106]. HxA3-C is converted to HxA3-D by γ-glutamyltranspeptidase. HxA3 and HxB3 can also be converted into trioxilin A3 (TrXA3) (8,11,12-trihydroxyepoxyeicosatrienoic acid) and TrXB3 (10,11,12-trihydroxyepoxyeicosatrienoic acid) with soluble epoxide hydrolase (sEH) (current name: epoxide hydrolase 2 (EPHX2)) [103][107]. HxA3 receptors are TRPV1 and transient receptor potential ankyrin 1 (TRPA1) [108][109]. HxA3 and TrXA3 are also antagonists of the TP receptor [110], the receptor for TxA2.2.1.4. 12R-Lipoxygenase
In addition to 12S-LOX/ALOX12, there is a second enzyme with 12-LOX activity [13], namely 12R-LOX/ALOX12B [111]. This enzyme shows activity towards ARA C20:4n-6 but not linoleic acid C18:2n-6 [111]. 12R-LOX/ALOX12B transforms ARA C20:4n-6 into 12R-HpETE, a stereoisomer of the product of 12S-LOX/ALOX12’s enzyme activity. 12R-HpETE is converted to 11,12-bis-epi-HxA3 with eLOX3/ALOXE3 [18]. 12R-HpETE is a stereoisomer of 12S-HpETE. Similar to this compound, 12R-HpETE can also be converted to 12R-HETE [24], which is then converted to 12-oxo-ETE with an enzyme with 12-hydroxyeicosanoid dehydrogenase activity [24][87], including eLOX3/ALOXE3 [18]. The ALOX12B gene is only 38% similar to the ALOX12 gene. The highest expression of this enzyme is found in the skin, and it is much lower in the prostate and adrenal gland [14][17][111]. 12R-LOX is important in skin function; mutations in the ALOX12B gene lead to ichthyosis [26][28][112], as do mutations in the ALOXE3 gene. 12R-LOX/ALOX12B and eLOX3/ALOXE3 participate in a common pathway in lipid mediator production. 12R-LOX produces 12R-HpETE, which is converted to 11,12-bis-epi-HxA3 with eLOX3 (Figure 3) [18]. Under the influence of eLOX3/ALOXE3, 12-oxo-ETE is also formed from 12R-HpETE in small amounts [18].
Figure 3. 12-LOX pathway. ARA C20:4n-6 is converted to 12S-HpETE and 12R-HpETE with 12S-LOX and 12R-LOX, respectively. Either 12-oxo-ETE or the corresponding 12-HETE can be formed from these compounds. 12S-HpETE can also be converted to HxA3 or HxB3 with hemin and lipoxygenases: eLOX3, 12S-LOX, or 15-LOX-1. 12R-HpETE can undergo a similar conversion to 11,12-bis-epi-HxA3. HxA3 may undergo further transformations. HxA3 can be conjugated to glutathione. HxA3-C is then formed, from which amino acids can be detached—HxA3-D is then formed in a reaction similar to the transformation of cysteinyl-leukotrienes. HxA3 can also be converted to TrXA3. Arrows next to enzymes: higher or lower expression of given enzymes in GBM tumor relative to healthy tissue. ↓—lower expression of given enzymes in GBM tumor relative to healthy tissue.
2.1.5. 15-Lipoxygenases
Like the previously described LOX, 15-LOX catalyzes the formation of 15S-hydroperoxyeicosatetraenoic acids (15-HpETE) from ARA 20:4n-6 [113]. In humans, two 15-LOX isoforms are distinguished: 15-LOX-1/ALOX15 [114] and 15-LOX-2/ALOX15B [115]. The highest expression of 15-LOX-1/ALOX15 is found in the lung, and the lower expressions are in the skin, intestine, heart, lymph node, and testis [17]. The highest expression of 15-LOX-2/ALOX15B is found in the prostate and skin. Expression of this enzyme is also observed in the lung, esophagus, and cornea [14][17][115]. The enzymatic properties of the two isoforms differ. 15-LOX-1/ALOX15 catalyzes the formation of 15-HpETE, but it also converts part of the substrate, ARA 20:4n-6, into 12-HpETE [93]—for this reason, the enzyme owns its historical name: 12/15-LOX. 15-LOX-2/ALOX15B has no such activity [93][115]. 15-LOX-1/ALOX15 shows much higher activity with linoleic acid C18:2n-6 than 15-LOX-2/ALOX15B (Figure 4) [93]. These enzymes convert linoleic acid C18:2n-6 into 13S-hydroperoxyoctadecadienoic acid (13-HpODE), which converts to 13S-hydroxyoctadecadienoic acid (13-HODE). The identified receptor for 13-HpODE is G2A/GPR132 [35]. 13-HODE also activates the TRPV1 receptor [116]. 13-HODE undergoes the same transformations as HETE and can be oxidized to 13-oxo-ODE. 13-oxo-ODE [117] and 13-HODE [118] are PPARγ ligands.
Figure 4. 15-LOX pathway. (A). Linoleic acid C18:2n-6 can be converted by 15-LOX-1 and 15-LOX-2 into 13-HpODE. This compound can then be converted into 13-HODE and 13-oxo-ODE. (B) 15-LOX-1 and 15-LOX-2 can convert ARA C20:4n-6 into 15-HpETE. 15-LOX-1 can also convert this fatty acid into 12-HpETE. 15-HpETE can then be converted into EXA4 and into cysteinyl-eoxins EXC4, EXD4, and EXE4. 15-HpETE can also be converted into hepoxilins 14,15-HxA3 11S, and 14,15-HxB3 13R. 14,15-HxA3 11S can be converted to cysteinyl hepoxilins, such as 14,15-HxA3-C 11S.
2.2. Lipoxygenases in Glioblastoma Multiforme
In GBM tumors, ARA C20:4n-6 is mainly processed by COX, as shown by experiments on the C6 cell line [128]. In contrast, in the healthy brain, this PUFA is mainly processed by the LOX pathway. This shows that in GBM tumors, the LOX pathway may not be as important as the COX pathway, although it is still important in tumor mechanisms in GBM tumors.2.2.1. 5-Lipoxygenase Pathway in Glioblastoma Multiforme
The expression of 5-LOX/ALOX5 in a GBM tumor is higher than in non-tumor brain tissue [129][130][131]. This is also confirmed by data obtained from the GEPIA portal [8] and from Seifert et al. transcriptomics analysis [9]. Expression of 5-LOX/ALOX5 in the GBM tumor is found in macrophage and microglial cells as well as in other cells, such as cancer cells [130][131]. It is higher in GBM cancer stem cells than in other GBM cancer cells [132]. According to GEPIA, higher expressions of FLAP/ALOX5AP, LTC4S, LTA4H, GGT5, and DPEP1 but not DPEP2 [8], the enzymes that synthesize LTB4 and LTE4 from the product of 5-LOX/ALOX5 activity, were also found in GBM tumors [43][44][49][50][52][54]. Seifert et al. showed that there are higher expressions of FLAP/ALOX5AP, LTA4H, and GGT5 in GBM tumors than in healthy brain tissue [9]. In contrast, LTC4S, DPEP1, and DPEP2 are not affected. The higher expression of enzymes responsible for leukotriene biosynthesis increases the production [133] and levels [134] of these lipid mediators further in GBM tumors than in healthy brain tissue, particularly cysteinyl-leukotrienes. The expression level of 5-LOX/ALOX5 in GBM tumors does not affect prognosis [8][135], although simultaneous high expression of COX-2 and 5-LOX/ALOX5, two major ARA C20:4n-6 processing enzymes, is associated with a worse prognosis [135]. This shows that the two pathways in cooperation can impinge on prognosis severity. The expression levels of most enzymes involved in leukotriene production and metabolism do not affect prognosis [8]. Only for GGT1, higher expression in GBM tumors is associated with a worse prognosis [8]. GGT5 expression showed a positive trend (p = 0.055) toward a worse prognosis. GGT1 and GGT5 are enzymes that catalyze the transformation of LTC4 into LTD4 [52], demonstrating that the transformation of cysteinyl leukotrienes may be important in tumorigenesis in GBM. In addition, higher expression of 12-HEDH/PTGR1, an enzyme that degrades LTB4, as well as prostaglandins, may be associated with worse prognoses for GBM patients [86], although GEPIA did not confirm such a link [8]. In addition, GEPIA and Seifert et al. did not show that 12-HEDH/PTGR1 expression differs between GBM tumors and healthy brain tissue [8][9]. According to GEPIA [8] and Seifert et al. [9], expression levels of receptors for leukotrienes LTB4R1, LTB4R2, CysLTR1, CysLTR2, GPR17, and OXGR1/GPR99 do not differ between GBM tumors and healthy brain tissue. In addition, the expression levels of these receptors in GBM tumors do not affect prognosis [8]. Leukotrienes as well as the entire 5-LOX pathway are important in tumorigenesis in GBM. They may also be important in the onset of GBM and in the first stages of tumorigenesis. The GA genotype of rs2291427 in the ALOX5 gene is associated with a higher risk of GBM in men [136]. Expression of 5-LOX/ALOX5 is higher in GBM cancer stem cells than in other GBM cancer cells [132]. The products of 5-LOX/ALOX5 activity induce proliferation and self-renewal of GBM cancer stem cells. The effects of 5-LOX/ALOX5 on GBM cancer stem cells are autocrine in nature. LTB4 also increases the proliferation of GBM cells [137]. This is associated with an increase in Ca2+ levels in the cytoplasm of GBM cells [137]. Studies of various cell lines show that 5-LOX/ALOX5 expression is present in only a portion of them [138][139]. Expression of 5-LOX/ALOX5 causes an autocrine increase in the proliferation of such a line and, thus, makes culture growth dependent on 5-LOX/ALOX5 activity. All GBM lines express LTA4H, LTB4R1/BLT1, LTB4R2/BLT2, and CysLTR2, but only some lines express LTC4S [139], indicating heterogeneity in the production of cysteinyl-leukotrienes and 5-HETE by GBM cancer cells. The dependence of the proliferation of some GBM cancer cell lines on the 5-LOX pathway may be a potential therapeutic target for GBM treatment in personalized therapy. For this reason, the pan-LOX inhibitor Nordy [132][140], 5-LOX inhibitors such as caffeic acid [137], A861 [141], AA-863, and U-60,257 (pyriprost) [142], LTA4H inhibitors such as bestatin [141], and CysLTR1 and CysLTR2 receptor inhibitors such as montelukast and zafirlukast [143] have anti-tumor properties against GBM and inhibit proliferation. This is associated with decreased ERK MAPK activation and induction of apoptosis as a result of decreased expression of anti-apoptotic Bcl-2 and increased expression of pro-apoptotic Bax [138]. Cysteinyl leukotrienes may have anticancer properties by increasing the bioavailability of various chemotherapeutics. In the brain, as well as in GBM tumors, there is a blood-brain barrier (BBB) that is poorly permeable to many substances, including anticancer drugs [144]. However, cysteinyl leukotrienes have BBB permeability, as shown by experiments on rat RG-2 glioma tumors [145]. BBB permeability is highest for LTE4 [145], with cysteinyl leukotrienes not causing BBB permeability in healthy brain tissue [145][146]. For this reason, the administration of LTC4 prior to the administration of chemotherapeutics that pass poorly through the BBB increases the bioavailability of drugs such as cisplatin [147]. However, this method does not increase the bioavailability of all chemotherapeutics, as exemplified by paclitaxel [148]. The receptor for cysteinyl leukotrienes is GPR17 [79]. According to GEPIA [8] and Seifert et al. [9], the expression level of this receptor does not differ between GBM tumors and healthy brain tissue. Higher GPR17 expression is associated with better prognosis in patients with low-grade gliomas, according to the Chinese Glioma Genome Atlas (CGGA) [149] and GEPIA [8], but the expression of this receptor is not associated with prognosis in a GBM patient [8]. GPR17 expression is also higher in low-grade gliomas than in healthy brain tissue [149]. Activation of this receptor by the ligand inhibits proliferation in the G1 phase and induces apoptosis of GBM cell lines LN-229 and SNB-19 [149]. In addition, GPR17 ligands inhibit tumor growth, as shown by experiments using patient-derived xenograft mouse models. The action of GPR17 is associated with a decrease in the levels of cyclic adenosine monophosphate (cAMP) and Ca2+ in the cytoplasm, which reduces the activation of the PI3K → Akt/PKB pathway [149][150]. An increase in GPR17 expression can cause the proliferation and migration of GBM cells [151], particularly with an increase in the expression of this receptor by long non-coding RNA (lncRNA) colorectal neoplasia differentially expressed (CRNDE) in low-grade glioma cells [151]. The receptor for 5-HETE, and also other lipid mediators, is G2A/GPR132 [35]. Higher expression of this receptor, according to GEPIA, is associated with a worse prognosis for a GBM patient (p = 0.052) [8], yet there is no significant upregulation of this receptor expression in GBM tumors [8][9]. 5-oxo-ETE may also play an important role in tumorigenic mechanisms in GBM. The receptor for this lipid mediator is OXER1/GPR99 [39][40][41]. The expression of this receptor does not differ between GBM tumor and healthy brain tissue [8][9]. According to GEPIA, higher expression of OXER1/GPR99, the receptor for 5-oxo-ETE, is associated with a worse prognosis for a GBM patient [8]. OXER1/GPR99 is also a receptor for 2-oxoglutarate, LTC4, and LTD4 [78]. There is a lack of thorough research on the importance of 5-oxo-ETE in tumorigenesis in GBM tumors.2.2.2. 12-Lipoxygenase Pathway in Glioblastoma Multiforme
In GBM tumors, expression of 12S-LOX/ALOX12 and 12R-LOX/ALOX12B is not different from healthy brain tissue [8][9], nor is it associated with prognosis severity [8], nor is the expression of the receptor for 12S-HETE, i.e., GPR31, elevated and affecting prognosis [8][9]. In contrast, the expression of eLOX3/ALOXE3 in GBM tumors is lower than in other brain tissue [8][23]. On the other hand, the transcriptomics analysis by Seifert et al. showed no differences between eLOX3/ALOXE3 expression levels in GBM tumor and healthy brain tissue [9]. Downregulation of eLOX3/ALOXE3 expression in GBM tumor is associated with increased expression of miR-18a, which downregulates eLOX3/ALOXE3 expression [23]. At the same time, eLOX3/ALOXE3 expression is also not related to the prognoses of GBM patients [8]. 12-LOX is involved in tumorigenesis in GBM. Studies on various cell lines have shown that 12-LOX expression is common in GBM cancer cells [139]. For this reason, 12-LOX inhibitors inhibit proliferation and reduce the viability of GBM cells [139][152]. 12-LOX inhibitors also inhibit the migration of GBM cells because they reduce the expression of matrix metalloproteinase 2 (MMP2) in these cells [139]. However, the exact mechanism of 12-LOX action on tumorigenic processes in GBM is poorly studied. The fact that eLOX3/ALOXE3 is anticancer in nature [23] suggests that a lipid mediator not formed by eLOX3/ALOXE3 is responsible for the pro-cancer properties of 12-LOX. Perhaps it is 12-HETE, a lipid mediator with proven pro-cancer properties in other cancers [153][154]. In addition, higher expression of G2A/GPR132, a receptor for 5-HETE, 12-HETE, 15-HETE, and 9-HODE, is associated with a worse prognosis for a GBM patient (p = 0.052) [8]. The oncogenic properties of G2A/GPR132 were also demonstrated in a study on fibroblasts [155], although there is no higher expression of G2A/GPR132 in GBM tumors than in healthy brain tissue [8][9]. 12-LOX may also have anti-cancer properties. It converts ARA 20:4n-6 into 12-HpETE, a lipid from the hydroperoxyl group, and for this reason, it can cause lipid peroxidation, which, when free ARA 20:4n-6 is in excess and this PUFA is over-processed, has a destructive effect on the cell [156]. eLOX3/ALOXE3 has anti-tumor properties in GBM. eLOX3/ALOXE3 converts 12-HpETE into 12-oxo-ETE. In the absence of eLOX3/ALOXE3, 12-HpETE is converted to 12-HETE [23], meaning that eLOX3/ALOXE3 decreases 12-HETE production. This lipid mediator increases GBM cell migration. When 12-HETE production is decreased, GBM cell migration is reduced. The lipid mediators produced by eLOX3/ALOXE3, including 12-oxo-ETE, have anti-tumor effects, particularly 12-oxo-ETE, which is a ligand for PPARγ [22][23]. Activation of this nuclear receptor inhibits proliferation and induces apoptosis of GBM cancer cells [157][158][159]. The products of eLOX3/ALOXE3 activity are hepoxilins and trioxilins [18][21], lipid mediators of physiological importance. However, there is a lack of studies on the importance of these lipid mediators in tumorigenesis in GBM. Analysis on the GEPIA portal [8] and the transcriptomics analysis by Seifert et al. [9] showed no differences in the expression of EPHX2, the enzyme responsible for converting hepoxilins into trioxilins, between GBM tumors and healthy brain tissue [103][107]. At the same time, according to GEPIA, higher EPHX2 expression in GBM tumors is associated with a tendency toward a worse prognosis (p = 0.072), which may indicate that hepoxilins and trioxilins may have some role in neoplastic processes in GBM.2.2.3. 15-Lipoxygenase Pathway in Glioblastoma Multiforme
GEPIA [8] and Seifert et al. [9] showed no differences in the expression of 15-LOX-1/ALOX15 and 15-LOX-2/ALOX15B between GBM tumors and healthy brain tissue. According to GEPIA, the expression level of these enzymes does not affect the prognosis for patients [8]. Studies on various GBM lines have shown differences in the expression of 15-LOX-1/ALOX15 and 15-LOX-2/ALOX15B in GBM cancer cells [139]. 15-LOX is important in the function of GBM cancer cells, and 15-LOX inhibitors reduce the viability and migration of GBM cancer cells [139]. On the other hand, increasing the expression and activity of 15-LOX-1/ALOX15 throughout the body may have an anti-tumor effect against GBM, as shown by gene therapy using an adenovirus transducing the ALOX15 gene [160]. This effect may depend on 13-HODE and 15-HETE. All GBM lineages secrete 13-HODE, a product of the linoleic acid C18:2n-6 conversion with 15-LOX-1/ALOX15 and 15-LOX-2/ALOX15B [93]. 13-HODE increases MMP2 expression in GBM cells, which causes migration [139]. At the same time, 13-HODE also decreases the viability of GBM cells [139], which may depend on the activation of PPARγ via this lipid mediator [118]. This mechanism was confirmed in other cancers, including non-small cell lung cancer [161]. 15-HETE can activate G2A/GPR132 [35]. Higher expression of this receptor. according to GEPIA. is associated with a worse prognosis for a GBM patient (p = 0.052) [8]. At the same time, the importance of this receptor in GBM has not been thoroughly investigated. Studies in other models have shown that G2A/GPR132 is an oncogene [155]; that is, 15-HETE through activation of G2A/GPR132 has a pro-cancer effect. At the same time, there is no significant upregulation of this receptor expression in GBM tumors [8][9]. The significance of lipoxins in GBM tumors has not been thoroughly investigated. The expression level of the LXA4 receptor ALX/FPR2 does not differ between GBM tumors and healthy brain tissue (Table 1) [8][9]. The expression level of this receptor in GBM tumors does not affect prognosis. However, it may be important in tumorigenesis in GBM tumors. Studies on U-87 MG cells have shown that silencing ALX/FPR2 reduces the proliferation and migration of the cells tested [162]. In addition, cells with silenced ALX/FPR2 showed lower expressions of VEGF, a major pro-angiogenic factor. However, this receptor is activated not only by LXA4 but also by other factors [163]—for this reason, the importance of LXA4 in tumorigenic processes in GBM cannot be determined.Table 1. Description of the various enzymes involved in the synthesis, action, and degradation of lipoxygenases along with their involvement in tumorigenesis in GBM.
| Name | Biochemical Significance | Expression Level In GBM Tumors Relative To Healthy Tissue | Impact on Prognosis with Higher Expression in GBM Tumors | |
|---|---|---|---|---|
| Source | GEPIA [8] | Seifert et al. [9] | GEPIA [8] | |
| eLOX3/ALOXE3 | Production of hepoxilins/hydroxy-epoxyeicosatrienoic acid and oxo-ETE from HpETE | Lower expression in the tumor | Expression does not change | No significant impact on prognosis |
| 5-LOX/ALOX5 | 5-HpETE production from ARA; the first enzyme in leukotrienes and the 5-oxo-ETE synthesis pathway; synthesis of lipoxins from 15-HpETE and 15-HETE | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| FLAP/ALOX5AP | Substrate carrier for 5-LOX | Higher expression in the tumor | Higher expression in the tumor | No significant impact on prognosis |
| 12S-LOX/ALOX12 | 12S-HpETE production from ARA; the first enzyme in the hepoxilin production pathway; production of lipoxins from LTA4 | Expression does not change | Expression does not change | No significant impact on prognosis |
| 12R-LOX/ALOX12B | 12R-HpETE production from ARA | Expression does not change | Expression does not change | No significant impact on prognosis |
| 15-LOX-1/ALOX15 | 15-HpETE production from ARA; 12-HpETE production from ARA; production of lipoxins, eoxins, 15-oxo-ETE and 15-HETE; production of 13-HpODE from linoleic acid C18:2n-6 | Expression does not change | Expression does not change | No significant impact on prognosis |
| 15-LOX-2/ALOX15B | 15-HpETE production from ARA; production of 15-HpETE, lipoxins, eoxins, 15-oxo-ETE and 15-HETE | Expression does not change | Expression does not change | No significant impact on prognosis |
| LTA4H | LTB4 production from LTA4 | Higher expression in the tumor | Higher expression in the tumor | |
| GPR17 | ||||
| Cysteinyl-leukotrienes receptor | ||||
| Expression does not change | ||||
| Expression does not change | ||||
| No significant impact on prognosis | ||||
| GPR31 | ||||
| 12 | S-HETE receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| OXGR1/GPR99 | LTE4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| G2A/GPR132 | 5-HETE, 12-HETE, 15-HETE, 9-HODE receptor | Expression does not change | Expression does not change | Worse prognosis (p = 0.052) |
Red background—higher expression in the tumor; blue background—lower expression in the tumor; red background—worse prognosis with higher expression.
The expression levels of various LOX are not associated with prognoses for GBM patients [8]. This indicates that the LOX pathway is not as relevant to cancer processes as other pathways. For this reason, drugs targeting LOX may show poor efficacy in GBM therapy. At the same time, the analyses performed in this research show that higher expression of OXER1 (the receptor for 5-oxo-ETE) and higher expression of G2A/GPR132 (the receptor for various HETE) are associated with poor prognosis [8]. This indicates a therapeutic target for future drugs developed for the treatment of GBM. In addition, higher expression of GGT1 in GBM tumors is associated with worse prognosis, and higher expression of GGT5 and EPHX2 is associated with a trend of worse prognosis for GBM patients. This indicates a future direction for research into tumor mechanisms in GBM.
2.3. Pan-Cancer Analysis of Genes Related to LOX Pathway and GBM
Similar to the COX pathway, a pan-cancer analysis of the expression of the genes involved in the LOX pathway using the data from the GEPIA web server were performed [8]. The expression of eLOX3/LOXE3 is reduced in GBM tumors. At the same time, there is no change in the expression of this enzyme relative to healthy brain tissue in lower grade gliomas. It is also reduced in two more types of tumors. For this reason, a decrease in eLOX3/LOXE3 expression may be considered specific to GBM. In GBM tumors, there is elevated expression of 5-LOX/ALOX5 and FLAP/ALOX5AP relative to healthy brain tissue, which is similar to lower grade gliomas [8]. Expression of these proteins is elevated in 9 and 11 tumor types, respectively. In a similar number of tumor types, there is a reduction in the expressions of 5-LOX/ALOX5 and FLAP/ALOX5AP. This indicates that the elevated expressions of 5-LOX/ALOX5 and FLAP/ALOX5AP may be glioma-specific. The expression of other LOX is not altered in GBM and lower grade gliomas, which is similar to most other types of cancer. In GBM tumors, there are elevated expressions of LTA4H/LTA4H and LTC4S/LTC4S relative to healthy tissue [8]. In lower grade gliomas, there is higher expression of only LTC4S/LTC4S [8]. According to Seifert et al., in II and III grade gliomas, there are higher expressions of LTA4H/LTA4H but not LTC4S/LTC4S relative to healthy brain tissue [9]. LTA4H/LTA4H expression is elevated in 4 out of 31 analyzed tumor types. LTC4S/LTC4S is upregulated in six tumor types but downregulated in eleven types [8]. Therefore, the elevated expression of LTA4H/LTA4H and LTC4S/LTC4S can be considered as specific to GBM and glioma, respectively. GGT5 expression is upregulated in GBM and lower grade gliomas [8][9]. It is downregulated in eleven tumor types and upregulated in seven. Therefore, the elevation of GGT5 expression can be considered characteristic for gliomas. DPEP1 expression is elevated in GBM tumors but not in lower grade gliomas (Table 2) [8]. It is decreased in six types of tumors but increased in four types, including GBM. For this reason, it can be thought that changes in DPEP1 expression are characteristic of GBM. EPHX2 expression is often decreased in tumors. In a pan-cancer analysis, 17 types of tumors had a reduced expression of this enzyme relative to healthy tissue. At the same time, in GBM tumors, EPHX2 expression does not differ relative to healthy brain tissue [8][9].Table 2. Pan-cancer analysis of expression of genes involved in the LOX pathway.
| Name of Cancer | eLOX3/ALOXE3 | 5-LOX/ALOX5 | FLAP/ALOX5AP | 12S-LOX/ALOX12 | 12R-LOX/ALOX12B | 15-LOX-1/ALOX15 | 15-LOX-2/ALOX15B | LTA4H/LTA4H | LTC4S/LTC4S | GGT1 | GGT5 | DPEP1 | DPEP2 | EPHX2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | = | ↓ | ↓ | = | = | = | ↓ | = | = | = | ↓ | = | = | ↓ | |||||
| Bladder urothelial carcinoma (BLCA) | = | = | ↓ | = | = | = | = | = | ↓ | = | ↓ | = | = | ↓ | |||||
| Breast invasive carcinoma (BRCA) | = | = | = | = | = | = | ↓ | = | = | = | = | = | = | = | |||||
| Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC) | = | = | = | = | = | = | = | = | ↓ | = | ↓ | = | = | ↓ | |||||
| Cholangiocarcinoma (CHOL) | = | ↑ | ↑ | = | = | = | = | ↑ | ↑ | = | = | = | = | ↓ | |||||
| Colon adenocarcinoma (COAD) | = | = | = | = | = | = | = | = | ↓ | ↑ | = | ↑ | ↓ | = | |||||
| Lymphoid neoplasm diffuse large B-cell lymphoma (DLBC) | = | ↓ | ↓ | ↓ | = | = | ↑ | = | = | = | ↑ | = | ↓ | = | |||||
| Esophageal carcinoma (ESCA) | = | = | = | No significant impact on prognosis | |||||||||||||||
| ↓ | = | = | ↓ | = | = | = | = | = | = | ↓ | LTC4S | LTC4 production from LTA4 | Higher expression in the tumor | Expression does not change | No significant impact on prognosis | ||||
| Glioblastoma multiforme (GBM) | ↓ | ↑ | ↑ | = | = | = | = | ↑ | ↑ | = | ↑ | ↑ | = | = | GGT1 | LTD4 production from LTC4 | Expression does not change | Expression does not change | Worse prognosis |
| Head and neck squamous cell carcinoma (HNSC) | ↑ | = | = | ↓ | = | = | = | = | = | = | ↑ | = | = | ↓ | GGT5 | LTD4 production from LTC4 | |||
| Kidney chromophobe (KICH) | Higher expression in the tumor | Higher expression in the tumor | Worse prognosis ( | p = 0.055) | |||||||||||||||
| = | = | = | = | = | = | = | = | ↓ | ↓ | ↓ | ↓ | = | = | DPEP1 | LTE4 production from LTD4 | Higher expression in the tumor | Expression does not change | No significant impact on prognosis | |
| Kidney renal clear cell carcinoma (KIRC) | = | ↑ | ↑ | = | = | = | = | = | = | ↑ | = | ↓ | ↑ | ↓ | DPEP2 | LTE4 production from LTD4 | Expression does not change | Expression does not change | No significant impact on prognosis |
| Kidney renal papillary cell carcinoma (KIRP) | = | ↑ | ↑ | = | = | = | ↑ | = | = | ↑ | ↓ | ↓ | ↑ | = | EPHX2 | Conversion of hepoxilins into trioxilin | Expression does not change | Expression does not change | Worse prognosis (p = 0.072) |
| Acute myeloid leukemia (LAML) | = | ↑ | ↑ | = | = | = | = | ↑ | ↑ | = | ↑ | = | ↑ | ↓ | Receptors | ||||
| Brain lower grade glioma (LGG) | = | ↑ | ↑ | = | = | = | = | = | ↑ | = | ↑ | = | = | = | LTB4R1 | LTB4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| Liver hepatocellular carcinoma (LIHC) | = | = | = | = | = | = | = | = | = | = | ↓ | = | = | ↓ | LTB4R2 | LTB4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| CYSLTR1 | Cysteinyl-leukotrienes receptor | ||||||||||||||||||
| Lung adenocarcinoma (LUAD) | = | ↓ | ↓ | = | = | = | ↓ | = | ↓ | = | = | = | ↓ | = | Expression does not change | Expression does not change | No significant impact on prognosis | ||
| Lung squamous cell carcinoma (LUSC) | = | ↓ | ↓ | = | = | = | ↓ | ↓ | ↓ | ↓ | = | = | ↓ | = | CYSLTR2 | Cysteinyl-leukotrienes receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| Ovarian serous cystadenocarcinoma (OV) | = | ↑ | ↑ | = | = | = | = | = | = | = | ↓ | = | = | ↓ | OXER1 | 5-oxo-ETE receptor | Expression does not change | Expression does not change | Worse prognosis |
| Pancreatic adenocarcinoma (PAAD) | = | ↑ | ↑ | = | = | = | ↑ | ↑ | ↑ | ↑ | ↑ | ↓ | ↑ | ↓ | ALX/FPR2 | LXA4 receptor | Expression does not change | Expression does not change | No significant impact on prognosis |
| Pheochromocytoma and paraganglioma (PCPG) | = | = | ↓ | = | = | = | = | = | = | = | ↓ | = | = | ↓ | |||||
| Prostate adenocarcinoma (PRAD) | = | = | = | = | = | = | = | = | = | ↑ | = | = | = | = | |||||
| Rectum adenocarcinoma (READ) | = | = | = | = | = | = | = | = | ↓ | ↑ | = | ↑ | = | = | |||||
| Sarcoma (SARC) | = | = | = | = | = | = | = | = | = | ↓ | = | ↓ | = | ↓ | |||||
| Skin cutaneous melanoma (SKCM) | ↓ | ↓ | = | ↓ | ↓ | = | ↓ | = | ↓ | = | ↓ | = | = | ↓ | |||||
| Stomach adenocarcinoma (STAD) | = | = | ↑ | = | = | = | = | = | = | = | = | = | = | = | |||||
| Testicular germ cell tumors (TGCT) | ↓ | = | ↑ | = | = | ↓ | = | = | ↓ | = | = | ↓ | = | ↓ | |||||
| Thyroid carcinoma (THCA) | = | ↑ | ↑ | = | = | = | ↑ | = | = | = | = | = | = | = | |||||
| Thymoma (THYM) | = | ↓ | ↓ | = | = | = | = | = | ↑ | = | ↑ | ↑ | ↓ | ↑ | |||||
| Uterine corpus endometrial carcinoma (UCEC) | = | ↓ | = | = | = | = | = | = | ↓ | ↑ | ↓ | = | = | ↓ | |||||
| Uterine carcinosarcoma (UCS) | = | ↓ | ↓ | = | = | = | = | = | ↓ | = | ↓ | = | = | ↓ | |||||
Red background, ↑—expression higher in tumor than in healthy tissue; blue background, ↓—expression lower in tumor than in healthy tissue; gray background, =—expression does not differ between tumor and healthy tissue.
References
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199.
- Park, W.J.; Kothapalli, K.S.; Lawrence, P.; Tyburczy, C.; Brenna, J.T. An alternate pathway to long-chain polyunsaturates: The FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 2009, 50, 1195–1202.
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185.
- Leonard, A.E.; Bobik, E.G.; Dorado, J.; Kroeger, P.E.; Chuang, L.T.; Thurmond, J.M.; Parker-Barnes, J.M.; Das, T.; Huang, Y.S.; Mukerji, P. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 2000, 350 Pt 3, 765–770.
- Leonard, A.E.; Kelder, B.; Bobik, E.G.; Chuang, L.T.; Lewis, C.J.; Kopchick, J.J.; Mukerji, P.; Huang, Y.S. Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 2002, 37, 733–740.
- Kitazawa, H.; Miyamoto, Y.; Shimamura, K.; Nagumo, A.; Tokita, S. Development of a high-density assay for long-chain fatty acyl-CoA elongases. Lipids 2009, 44, 765–773.
- Shakya, S.; Gromovsky, A.D.; Hale, J.S.; Knudsen, A.M.; Prager, B.; Wallace, L.C.; Penalva, L.O.F.; Brown, H.A.; Kristensen, B.W.; Rich, J.N.; et al. Altered lipid metabolism marks glioblastoma stem and non-stem cells in separate tumor niches. Acta Neuropathol. Commun. 2021, 9, 101.
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102.
- Seifert, M.; Garbe, M.; Friedrich, B.; Mittelbronn, M.; Klink, B. Comparative transcriptomics reveals similarities and differences between astrocytoma grades. BMC Cancer 2015, 15, 952.
- Korbecki, J.; Kojder, K.; Jeżewski, D.; Simińska, D.; Tarnowski, M.; Kopytko, P.; Safranow, K.; Gutowska, I.; Goschorska, M.; Kolasa-Wołosiuk, A.; et al. Expression of SCD and FADS2 Is Lower in the Necrotic Core and Growing Tumor Area than in the Peritumoral Area of Glioblastoma Multiforme. Biomolecules 2020, 10, 727.
- Korbecki, J.; Simińska, D.; Jeżewski, D.; Kojder, K.; Tomasiak, P.; Tarnowski, M.; Chlubek, D.; Baranowska-Bosiacka, I. Glioblastoma Multiforme Tumors in Women Have a Lower Expression of Fatty Acid Elongases ELOVL2, ELOVL5, ELOVL6, and ELOVL7 than in Men. Brain Sci. 2022, 12, 1356.
- Jalota, A.; Kumar, M.; Das, B.C.; Yadav, A.K.; Chosdol, K.; Sinha, S. A drug combination targeting hypoxia induced chemoresistance and stemness in glioma cells. Oncotarget 2018, 9, 18351–18366.
- Funk, C.D.; Funk, L.B.; FitzGerald, G.A.; Samuelsson, B. Characterization of human 12-lipoxygenase genes. Proc. Natl. Acad. Sci. USA 1992, 89, 3962–3966.
- Krieg, P.; Marks, F.; Fürstenberger, G. A gene cluster encoding human epidermis-type lipoxygenases at chromosome 17p13.1: Cloning, physical mapping, and expression. Genomics 2001, 73, 323–330.
- Jisaka, M.; Kim, R.B.; Boeglin, W.E.; Brash, A.R. Identification of amino acid determinants of the positional specificity of mouse 8S-lipoxygenase and human 15S-lipoxygenase-2. J. Biol. Chem. 2000, 275, 1287–1293.
- Fürstenberger, G.; Marks, F.; Krieg, P. Arachidonate 8(S)-lipoxygenase. Prostaglandins Other Lipid Mediat. 2002, 68–69, 235–243.
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 2014, 13, 397–406.
- Yu, Z.; Schneider, C.; Boeglin, W.E.; Marnett, L.J.; Brash, A.R. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc. Natl. Acad. Sci. USA 2003, 100, 9162–9167.
- Zheng, Y.; Brash, A.R. Dioxygenase activity of epidermal lipoxygenase-3 unveiled: Typical and atypical features of its catalytic activity with natural and synthetic polyunsaturated fatty acids. J. Biol. Chem. 2010, 285, 39866–39875.
- Yu, Z.; Schneider, C.; Boeglin, W.E.; Brash, A.R. Human and mouse eLOX3 have distinct substrate specificities: Implications for their linkage with lipoxygenases in skin. Arch. Biochem. Biophys. 2006, 455, 188–196.
- Gregus, A.M.; Dumlao, D.S.; Wei, S.C.; Norris, P.C.; Catella, L.C.; Meyerstein, F.G.; Buczynski, M.W.; Steinauer, J.J.; Fitzsimmons, B.L.; Yaksh, T.L.; et al. Systematic analysis of rat 12/15-lipoxygenase enzymes reveals critical role for spinal eLOX3 hepoxilin synthase activity in inflammatory hyperalgesia. FASEB J. 2013, 27, 1939–1949.
- Higgins, C.B.; Zhang, Y.; Mayer, A.L.; Fujiwara, H.; Stothard, A.I.; Graham, M.J.; Swarts, B.M.; DeBosch, B.J. Hepatocyte ALOXE3 is induced during adaptive fasting and enhances insulin sensitivity by activating hepatic PPARγ. JCI Insight 2018, 3, e120794.
- Yang, X.; Liu, J.; Wang, C.; Cheng, K.K.; Xu, H.; Li, Q.; Hua, T.; Jiang, X.; Sheng, L.; Mao, J.; et al. miR-18a promotes glioblastoma development by down-regulating ALOXE3-mediated ferroptotic and anti-migration activities. Oncogenesis 2021, 10, 15.
- Powell, W.S.; Rokach, J. Biosynthesis, biological effects, and receptors of hydroxyeicosatetraenoic acids (HETEs) and oxoeicosatetraenoic acids (oxo-ETEs) derived from arachidonic acid. Biochim. Biophys. Acta 2015, 1851, 340–355.
- Pace-Asciak, C.R. Pathophysiology of the hepoxilins. Biochim. Biophys. Acta 2015, 1851, 383–396.
- Yu, Z.; Schneider, C.; Boeglin, W.E.; Brash, A.R. Mutations associated with a congenital form of ichthyosis (NCIE) inactivate the epidermal lipoxygenases 12R-LOX and eLOX3. Biochim. Biophys. Acta 2005, 1686, 238–247.
- Wang, T.; Xu, C.; Zhou, X.; Li, C.; Zhang, H.; Lian, B.Q.; Lee, J.J.; Shen, J.; Liu, Y.; Lian, C.G. Homozygous ALOXE3 Nonsense Variant Identified in a Patient with Non-Bullous Congenital Ichthyosiform Erythroderma Complicated by Superimposed Bullous Majocchi’s Granuloma: The Consequences of Skin Barrier Dysfunction. Int. J. Mol. Sci. 2015, 16, 21791–21801.
- Hotz, A.; Kopp, J.; Bourrat, E.; Oji, V.; Komlosi, K.; Giehl, K.; Bouadjar, B.; Bygum, A.; Tantcheva-Poor, I.; Hellström Pigg, M.; et al. Meta-Analysis of Mutations in ALOX12B or ALOXE3 Identified in a Large Cohort of 224 Patients. Genes 2021, 12, 80.
- Ueda, N.; Kaneko, S.; Yoshimoto, T.; Yamamoto, S. Purification of arachidonate 5-lipoxygenase from porcine leukocytes and its reactivity with hydroperoxyeicosatetraenoic acids. J. Biol. Chem. 1986, 261, 7982–7988.
- Mancini, J.A.; Abramovitz, M.; Cox, M.E.; Wong, E.; Charleson, S.; Perrier, H.; Wang, Z.; Prasit, P.; Vickers, P.J. 5-lipoxygenase-activating protein is an arachidonate binding protein. FEBS Lett. 1993, 318, 277–281.
- Häfner, A.K.; Gerstmeier, J.; Hörnig, M.; George, S.; Ball, A.K.; Schröder, M.; Garscha, U.; Werz, O.; Steinhilber, D. Characterization of the interaction of human 5-lipoxygenase with its activating protein FLAP. Biochim. Biophys. Acta 2015, 1851, 1465–1472.
- Narala, V.R.; Adapala, R.K.; Suresh, M.V.; Brock, T.G.; Peters-Golden, M.; Reddy, R.C. Leukotriene B4 is a physiologically relevant endogenous peroxisome proliferator-activated receptor-alpha agonist. J. Biol. Chem. 2010, 285, 22067–22074.
- Falgueyret, J.; Riendeau, D. LTA4-derived 5-oxo-eicosatetraenoic acid: pH-dependent formation and interaction with the LTB4 receptor of human polymorphonuclear leukocytes. Biochim. Biophys. Acta 2000, 1484, 51–58.
- Chiba, N.; Imai, H.; Narashima, K.; Arai, M.; Oshima, G.; Kunimoto, M.; Nakagawa, Y. Cellular glutathione peroxidase as a predominant scavenger of hydroperoxyeicosatetraenoic acids in rabbit alveolar macrophages. Biol. Pharm. Bull. 1999, 22, 1047–1051.
- Obinata, H.; Hattori, T.; Nakane, S.; Tatei, K.; Izumi, T. Identification of 9-hydroxyoctadecadienoic acid and other oxidized free fatty acids as ligands of the G protein-coupled receptor G2A. J. Biol. Chem. 2005, 280, 40676–40683.
- Erlemann, K.R.; Cossette, C.; Grant, G.E.; Lee, G.J.; Patel, P.; Rokach, J.; Powell, W.S. Regulation of 5-hydroxyeicosanoid dehydrogenase activity in monocytic cells. Biochem. J. 2007, 403, 157–165.
- Patel, P.; Cossette, C.; Anumolu, J.R.; Erlemann, K.R.; Grant, G.E.; Rokach, J.; Powell, W.S. Substrate selectivity of 5-hydroxyeicosanoid dehydrogenase and its inhibition by 5-hydroxy-Delta6-long-chain fatty acids. J. Pharmacol. Exp. Ther. 2009, 329, 335–341.
- Nagendra Reddy, C.; Ye, Q.; Patel, P.; Sivendran, S.; Chourey, S.; Wang, R.; Anumolu, J.R.; Grant, G.E.; Powell, W.S.; Rokach, J. Design and synthesis of affinity chromatography ligands for the purification of 5-hydroxyeicosanoid dehydrogenase. Bioorg. Med. Chem. 2017, 25, 116–125.
- Hosoi, T.; Koguchi, Y.; Sugikawa, E.; Chikada, A.; Ogawa, K.; Tsuda, N.; Suto, N.; Tsunoda, S.; Taniguchi, T.; Ohnuki, T. Identification of a novel human eicosanoid receptor coupled to Gi/o. J. Biol. Chem. 2002, 277, 31459–31465.
- Jones, C.E.; Holden, S.; Tenaillon, L.; Bhatia, U.; Seuwen, K.; Tranter, P.; Turner, J.; Kettle, R.; Bouhelal, R.; Charlton, S.; et al. Expression and characterization of a 5-oxo-6E,8Z,11Z,14Z-eicosatetraenoic acid receptor highly expressed on human eosinophils and neutrophils. Mol. Pharmacol. 2003, 63, 471–477.
- Kalyvianaki, K.; Drosou, I.; Notas, G.; Castanas, E.; Kampa, M. Enhanced OXER1 expression is indispensable for human cancer cell migration. Biochem. Biophys. Res. Commun. 2021, 584, 95–100.
- Serhan, C.N. Lipoxin biosynthesis and its impact in inflammatory and vascular events. Biochim. Biophys. Acta 1994, 1212, 1–25.
- Rådmark, O.; Shimizu, T.; Jörnvall, H.; Samuelsson, B. Leukotriene A4 hydrolase in human leukocytes. Purification and properties. J. Biol. Chem. 1984, 259, 12339–12345.
- Rudberg, P.C.; Tholander, F.; Andberg, M.; Thunnissen, M.M.; Haeggström, J.Z. Leukotriene A4 hydrolase: Identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates. J. Biol. Chem. 2004, 279, 27376–27382.
- Paige, M.; Wang, K.; Burdick, M.; Park, S.; Cha, J.; Jeffery, E.; Sherman, N.; Shim, Y.M. Role of leukotriene A4 hydrolase aminopeptidase in the pathogenesis of emphysema. J. Immunol. 2014, 192, 5059–5068.
- Yokomizo, T.; Izumi, T.; Chang, K.; Takuwa, Y.; Shimizu, T. A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature 1997, 387, 620–624.
- Yokomizo, T.; Kato, K.; Terawaki, K.; Izumi, T.; Shimizu, T. A second leukotriene B4 receptor, BLT2. A new therapeutic target in inflammation and immunological disorders. J. Exp. Med. 2000, 192, 421–432.
- Krey, G.; Braissant, O.; L’Horset, F.; Kalkhoven, E.; Perroud, M.; Parker, M.G.; Wahli, W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997, 11, 779–791.
- Lam, B.K.; Penrose, J.F.; Freeman, G.J.; Austen, K.F. Expression cloning of a cDNA for human leukotriene C4 synthase, an integral membrane protein conjugating reduced glutathione to leukotriene A4. Proc. Natl. Acad. Sci. USA 1994, 91, 7663–7667.
- Ago, H.; Kanaoka, Y.; Irikura, D.; Lam, B.K.; Shimamura, T.; Austen, K.F.; Miyano, M. Crystal structure of a human membrane protein involved in cysteinyl leukotriene biosynthesis. Nature 2007, 448, 609–612.
- Strid, T.; Svartz, J.; Franck, N.; Hallin, E.; Ingelsson, B.; Söderström, M.; Hammarström, S. Distinct parts of leukotriene C4 synthase interact with 5-lipoxygenase and 5-lipoxygenase activating protein. Biochem. Biophys. Res. Commun. 2009, 381, 518–522.
- Wickham, S.; West, M.B.; Cook, P.F.; Hanigan, M.H. Gamma-glutamyl compounds: Substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal. Biochem. 2011, 414, 208–214.
- Adachi, H.; Kubota, I.; Okamura, N.; Iwata, H.; Tsujimoto, M.; Nakazato, H.; Nishihara, T.; Noguchi, T. Purification and characterization of human microsomal dipeptidase. J. Biochem. 1989, 105, 957–961.
- Habib, G.M.; Shi, Z.Z.; Cuevas, A.A.; Lieberman, M.W. Identification of two additional members of the membrane-bound dipeptidase family. FASEB J. 2003, 17, 1313–1315.
- Reddanna, P.; Prabhu, K.S.; Whelan, J.; Reddy, C.C. Carboxypeptidase A-catalyzed direct conversion of leukotriene C4 to leukotriene F4. Arch. Biochem. Biophys. 2003, 413, 158–163.
- Bernström, K.; Hammarström, S. A novel leukotriene formed by transpeptidation of leukotriene E. Biochem. Biophys. Res. Commun. 1982, 109, 800–804.
- Jedlitschky, G.; Keppler, D. Transport of leukotriene C4 and structurally related conjugates. Vitam. Horm. 2002, 64, 153–184.
- Zhang, D.W.; Nunoya, K.; Vasa, M.; Gu, H.M.; Cole, S.P.; Deeley, R.G. Mutational analysis of polar amino acid residues within predicted transmembrane helices 10 and 16 of multidrug resistance protein 1 (ABCC1): Effect on substrate specificity. Drug Metab. Dispos. 2006, 34, 539–546.
- Johnson, Z.L.; Chen, J. Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell 2017, 168, 1075–1085.
- Cui, Y.; König, J.; Buchholz, J.K.; Spring, H.; Leier, I.; Keppler, D. Drug resistance and ATP-dependent conjugate transport mediated by the apical multidrug resistance protein, MRP2, permanently expressed in human and canine cells. Mol. Pharmacol. 1999, 55, 929–937.
- Kamisako, T.; Leier, I.; Cui, Y.; König, J.; Buchholz, U.; Hummel-Eisenbeiss, J.; Keppler, D. Transport of monoglucuronosyl and bisglucuronosyl bilirubin by recombinant human and rat multidrug resistance protein 2. Hepatology 1999, 30, 485–490.
- Lee, Y.M.; Cui, Y.; König, J.; Risch, A.; Jäger, B.; Drings, P.; Bartsch, H.; Keppler, D.; Nies, A.T. Identification and functional characterization of the natural variant MRP3-Arg1297His of human multidrug resistance protein 3 (MRP3/ABCC3). Pharmacogenetics 2004, 14, 213–223.
- Rius, M.; Hummel-Eisenbeiss, J.; Keppler, D. ATP-dependent transport of leukotrienes B4 and C4 by the multidrug resistance protein ABCC4 (MRP4). J. Pharmacol. Exp. Ther. 2008, 324, 86–94.
- Belinsky, M.G.; Chen, Z.S.; Shchaveleva, I.; Zeng, H.; Kruh, G.D. Characterization of the drug resistance and transport properties of multidrug resistance protein 6 (MRP6, ABCC6). Cancer Res. 2002, 62, 6172–6177.
- Chen, Z.S.; Hopper-Borge, E.; Belinsky, M.G.; Shchaveleva, I.; Kotova, E.; Kruh, G.D. Characterization of the transport properties of human multidrug resistance protein 7 (MRP7, ABCC10). Mol. Pharmacol. 2003, 63, 351–358.
- Chen, Z.S.; Guo, Y.; Belinsky, M.G.; Kotova, E.; Kruh, G.D. Transport of bile acids, sulfated steroids, estradiol 17-beta-D-glucuronide, and leukotriene C4 by human multidrug resistance protein 8 (ABCC11). Mol. Pharmacol. 2005, 67, 545–557.
- Cattori, V.; van Montfoort, J.E.; Stieger, B.; Landmann, L.; Meijer, D.K.; Winterhalter, K.H.; Meier, P.J.; Hagenbuch, B. Localization of organic anion transporting polypeptide 4 (Oatp4) in rat liver and comparison of its substrate specificity with Oatp1, Oatp2 and Oatp3. Pflugers Arch. 2001, 443, 188–195.
- Li, L.; Lee, T.K.; Meier, P.J.; Ballatori, N. Identification of glutathione as a driving force and leukotriene C4 as a substrate for oatp1, the hepatic sinusoidal organic solute transporter. J. Biol. Chem. 1998, 273, 16184–16191.
- Yokomizo, T.; Kato, K.; Hagiya, H.; Izumi, T.; Shimizu, T. Hydroxyeicosanoids bind to and activate the low affinity leukotriene B4 receptor, BLT2. J. Biol. Chem. 2001, 276, 12454–12459.
- Okuno, T.; Iizuka, Y.; Okazaki, H.; Yokomizo, T.; Taguchi, R.; Shimizu, T. 12(S)-Hydroxyheptadeca-5Z, 8E, 10E-trienoic acid is a natural ligand for leukotriene B4 receptor 2. J. Exp. Med. 2008, 205, 759–766.
- Liu, M.; Saeki, K.; Matsunobu, T.; Okuno, T.; Koga, T.; Sugimoto, Y.; Yokoyama, C.; Nakamizo, S.; Kabashima, K.; Narumiya, S.; et al. 12-Hydroxyheptadecatrienoic acid promotes epidermal wound healing by accelerating keratinocyte migration via the BLT2 receptor. J. Exp. Med. 2014, 211, 1063–1078.
- Giusti, F.; Casiraghi, M.; Point, E.; Damian, M.; Rieger, J.; Bon, C.L.; Pozza, A.; Moncoq, K.; Banères, J.L.; Catoire, L.J. Structure of the agonist 12-HHT in its BLT2 receptor-bound state. Sci. Rep. 2020, 10, 2630.
- Hsu, P.Y.; Tsai, A.L.; Kulmacz, R.J.; Wang, L.H. Expression, purification, and spectroscopic characterization of human thromboxane synthase. J. Biol. Chem. 1999, 274, 762–769.
- Matsunobu, T.; Okuno, T.; Yokoyama, C.; Yokomizo, T. Thromboxane A synthase-independent production of 12-hydroxyheptadecatrienoic acid, a BLT2 ligand. J. Lipid Res. 2013, 54, 2979–2987.
- Lynch, K.R.; O’Neill, G.P.; Liu, Q.; Im, D.S.; Sawyer, N.; Metters, K.M.; Coulombe, N.; Abramovitz, M.; Figueroa, D.J.; Zeng, Z.; et al. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature 1999, 399, 789–793.
- Heise, C.E.; O’Dowd, B.F.; Figueroa, D.J.; Sawyer, N.; Nguyen, T.; Im, D.S.; Stocco, R.; Bellefeuille, J.N.; Abramovitz, M.; Cheng, R.; et al. Characterization of the human cysteinyl leukotriene 2 receptor. J. Biol. Chem. 2000, 275, 30531–30536.
- Nothacker, H.P.; Wang, Z.; Zhu, Y.; Reinscheid, R.K.; Lin, S.H.; Civelli, O. Molecular cloning and characterization of a second human cysteinyl leukotriene receptor: Discovery of a subtype selective agonist. Mol. Pharmacol. 2000, 58, 1601–1608.
- Kanaoka, Y.; Maekawa, A.; Austen, K.F. Identification of GPR99 protein as a potential third cysteinyl leukotriene receptor with a preference for leukotriene E4 ligand. J. Biol. Chem. 2013, 288, 10967–10972.
- Ciana, P.; Fumagalli, M.; Trincavelli, M.L.; Verderio, C.; Rosa, P.; Lecca, D.; Ferrario, S.; Parravicini, C.; Capra, V.; Gelosa, P.; et al. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. EMBO J. 2006, 25, 4615–4627.
- Qi, A.D.; Harden, T.K.; Nicholas, R.A. Is GPR17 a P2Y/leukotriene receptor? examination of uracil nucleotides, nucleotide sugars, and cysteinyl leukotrienes as agonists of GPR17. J. Pharmacol. Exp. Ther. 2013, 347, 38–46.
- Simon, K.; Merten, N.; Schröder, R.; Hennen, S.; Preis, P.; Schmitt, N.K.; Peters, L.; Schrage, R.; Vermeiren, C.; Gillard, M.; et al. The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes. Mol. Pharmacol. 2017, 91, 518–532.
- Maekawa, A.; Balestrieri, B.; Austen, K.F.; Kanaoka, Y. GPR17 is a negative regulator of the cysteinyl leukotriene 1 receptor response to leukotriene D4. Proc. Natl. Acad. Sci. USA 2009, 106, 11685–11690.
- Wheelan, P.; Zirrolli, J.A.; Morelli, J.G.; Murphy, R.C. Metabolism of leukotriene B4 by cultured human keratinocytes. Formation of glutathione conjugates and dihydro metabolites. J. Biol. Chem. 1993, 268, 25439–25448.
- Yokomizo, T.; Ogawa, Y.; Uozumi, N.; Kume, K.; Izumi, T.; Shimizu, T. cDNA cloning, expression, and mutagenesis study of leukotriene B4 12-hydroxydehydrogenase. J. Biol. Chem. 1996, 271, 2844–2850.
- Tobin, D.M.; Roca, F.J.; Ray, J.P.; Ko, D.C.; Ramakrishnan, L. An enzyme that inactivates the inflammatory mediator leukotriene B4 restricts mycobacterial infection. PLoS ONE 2013, 8, e67828.
- Panagopoulos, A.T.; Gomes, R.N.; Almeida, F.G.; da Costa Souza, F.; Veiga, J.C.E.; Nicolaou, A.; Colquhoun, A. The prostanoid pathway contains potential prognostic markers for glioblastoma. Prostaglandins Other Lipid Mediat. 2018, 137, 52–62.
- Wainwright, S.L.; Powell, W.S. Mechanism for the formation of dihydro metabolites of 12-hydroxyeicosanoids. Conversion of leukotriene B4 and 12-hydroxy-5,8,10,14-eicosatetraenoic acid to 12-oxo intermediates. J. Biol. Chem. 1991, 266, 20899–20906.
- Hagmann, W.; Korte, M. Hepatic uptake and metabolic disposition of leukotriene B4 in rats. Biochem. J. 1990, 267, 467–470.
- Berry, K.A.; Borgeat, P.; Gosselin, J.; Flamand, L.; Murphy, R.C. Urinary metabolites of leukotriene B4 in the human subject. J. Biol. Chem. 2003, 278, 24449–24460.
- Wainwright, S.; Falck, J.R.; Yadagiri, P.; Powell, W.S. Metabolism of 12(S)-hydroxy-5,8,10,14-eicosatetraenoic acid and other hydroxylated fatty acids by the reductase pathway in porcine polymorphonuclear leukocytes. Biochemistry 1990, 29, 10126–10135.
- Huber, M.; Müller, J.; Leier, I.; Jedlitschky, G.; Ball, H.A.; Moore, K.P.; Taylor, G.W.; Williams, R.; Keppler, D. Metabolism of cysteinyl leukotrienes in monkey and man. Eur. J. Biochem. 1990, 194, 309–315.
- Chang, W.C.; Ning, C.C.; Lin, M.T.; Huang, J.D. Epidermal growth factor enhances a microsomal 12-lipoxygenase activity in A431 cells. J. Biol. Chem. 1992, 267, 3657–3666.
- Wecksler, A.T.; Kenyon, V.; Deschamps, J.D.; Holman, T.R. Substrate specificity changes for human reticulocyte and epithelial 15-lipoxygenases reveal allosteric product regulation. Biochemistry 2008, 47, 7364–7375.
- Ikei, K.N.; Yeung, J.; Apopa, P.L.; Ceja, J.; Vesci, J.; Holman, T.R.; Holinstat, M. Investigations of human platelet-type 12-lipoxygenase: Role of lipoxygenase products in platelet activation. J. Lipid Res. 2012, 53, 2546–2559.
- Yeung, J.; Tourdot, B.E.; Adili, R.; Green, A.R.; Freedman, C.J.; Fernandez-Perez, P.; Yu, J.; Holman, T.R.; Holinstat, M. 12(S)-HETrE, a 12-Lipoxygenase Oxylipin of Dihomo-γ-Linolenic Acid, Inhibits Thrombosis via Gαs Signaling in Platelets. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 2068–2077.
- Guo, Y.; Zhang, W.; Giroux, C.; Cai, Y.; Ekambaram, P.; Dilly, A.K.; Hsu, A.; Zhou, S.; Maddipati, K.R.; Liu, J.; et al. Identification of the orphan G protein-coupled receptor GPR31 as a receptor for 12-(S)-hydroxyeicosatetraenoic acid. J. Biol. Chem. 2011, 286, 33832–33840.
- Sun, L.; Xu, Y.W.; Han, J.; Liang, H.; Wang, N.; Cheng, Y. 12/15-Lipoxygenase metabolites of arachidonic acid activate PPARγ: A possible neuroprotective effect in ischemic brain. J. Lipid Res. 2015, 56, 502–514.
- Falgueyret, J.P.; Leblanc, Y.; Riendeau, D. Stereoselective carbonyl reductases from rat skin and leukocyte microsomes converting 12-ketoeicosatetraenoic acid to 12(S)-HETE. FEBS Lett. 1990, 262, 197–200.
- Pace-Asciak, C.R. Hemoglobin- and hemin-catalyzed transformation of 12L-hydroperoxy-5,8,10,14-eicosatetraenoic acid. Biochim. Biophys. Acta 1984, 793, 485–488.
- Reynaud, D.; Demin, P.; Pace-Asciak, C.R. Hepoxilin A3 formation in the rat pineal gland selectively utilizes (12S)-hydroperoxyeicosatetraenoic acid (HPETE), but not (12R)-HPETE. J. Biol. Chem. 1994, 269, 23976–23980.
- Nigam, S.; Patabhiraman, S.; Ciccoli, R.; Ishdorj, G.; Schwarz, K.; Petrucev, B.; Kühn, H.; Haeggström, J.Z. The rat leukocyte-type 12-lipoxygenase exhibits an intrinsic hepoxilin A3 synthase activity. J. Biol. Chem. 2004, 279, 29023–29030.
- Krieg, P.; Rosenberger, S.; de Juanes, S.; Latzko, S.; Hou, J.; Dick, A.; Kloz, U.; van der Hoeven, F.; Hausser, I.; Esposito, I.; et al. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J. Investig. Dermatol. 2013, 133, 172–180.
- Pace-Asciak, C.R.; Lee, W.S. Purification of hepoxilin epoxide hydrolase from rat liver. J. Biol. Chem. 1989, 264, 9310–9313.
- Laneuville, O.; Chang, M.; Reddy, C.C.; Corey, E.J.; Pace-Asciak, C.R. Isozyme specificity in the conversion of hepoxilin A3 (HxA3) into a glutathionyl hepoxilin (HxA3-C) by the Yb2 subunit of rat liver glutathione S-transferase. J. Biol. Chem. 1990, 265, 21415–21418.
- Pace-Asciak, C.R.; Laneuville, O.; Chang, M.; Reddy, C.C.; Su, W.G.; Corey, E.J. New products in the hepoxilin pathway: Isolation of 11-glutathionyl hepoxilin A3 through reaction of hepoxilin A3 with glutathione S-transferase. Biochem. Biophys. Res. Commun. 1989, 163, 1230–1234.
- Pace-Asciak, C.R.; Laneuville, O.; Su, W.G.; Corey, E.J.; Gurevich, N.; Wu, P.; Carlen, P.L. A glutathione conjugate of hepoxilin A3: Formation and action in the rat central nervous system. Proc. Natl. Acad. Sci. USA 1990, 87, 3037–3041.
- Cronin, A.; Decker, M.; Arand, M. Mammalian soluble epoxide hydrolase is identical to liver hepoxilin hydrolase. J. Lipid Res. 2011, 52, 712–719.
- Gregus, A.M.; Doolen, S.; Dumlao, D.S.; Buczynski, M.W.; Takasusuki, T.; Fitzsimmons, B.L.; Hua, X.Y.; Taylor, B.K.; Dennis, E.A.; Yaksh, T.L. Spinal 12-lipoxygenase-derived hepoxilin A3 contributes to inflammatory hyperalgesia via activation of TRPV1 and TRPA1 receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 6721–6726.
- Singh, N.K.; Rao, G.N. Emerging role of 12/15-Lipoxygenase (ALOX15) in human pathologies. Prog. Lipid Res. 2019, 73, 28–45.
- Siangjong, L.; Goldman, D.H.; Kriska, T.; Gauthier, K.M.; Smyth, E.M.; Puli, N.; Kumar, G.; Falck, J.R.; Campbell, W.B. Vascular hepoxilin and trioxilins mediate vasorelaxation through TP receptor inhibition in mouse arteries. Acta Physiol. 2017, 219, 188–201.
- Boeglin, W.E.; Kim, R.B.; Brash, A.R. A 12R-lipoxygenase in human skin: Mechanistic evidence, molecular cloning, and expression. Proc. Natl. Acad. Sci. USA 1998, 95, 6744–6749.
- Zhang, L.; Hu, Y.; Lu, J.; Zhao, P.; Zhang, X.; Tan, L.; Li, J.; Xiao, C.; Zeng, L.; He, X. Identification of the first congenital ichthyosis case caused by a homozygous deletion in the ALOX12B gene due to chromosome 17 mixed uniparental disomy. Front. Genet. 2022, 13, 931833.
- Bryant, R.W.; Bailey, J.M.; Schewe, T.; Rapoport, S.M. Positional specificity of a reticulocyte lipoxygenase. Conversion of arachidonic acid to 15-S-hydroperoxy-eicosatetraenoic acid. J. Biol. Chem. 1982, 257, 6050–6055.
- Sigal, E.; Dicharry, S.; Highland, E.; Finkbeiner, W.E. Cloning of human airway 15-lipoxygenase: Identity to the reticulocyte enzyme and expression in epithelium. Am. J. Physiol. 1992, 262, L392–L398.
- Brash, A.R.; Boeglin, W.E.; Chang, M.S. Discovery of a second 15S-lipoxygenase in humans. Proc. Natl. Acad. Sci. USA 1997, 94, 6148–6152.
- Alsalem, M.; Wong, A.; Millns, P.; Arya, P.H.; Chan, M.S.; Bennett, A.; Barrett, D.A.; Chapman, V.; Kendall, D.A. The contribution of the endogenous TRPV1 ligands 9-HODE and 13-HODE to nociceptive processing and their role in peripheral inflammatory pain mechanisms. Br. J. Pharmacol. 2013, 168, 1961–1974.
- Altmann, R.; Hausmann, M.; Spöttl, T.; Gruber, M.; Bull, A.W.; Menzel, K.; Vogl, D.; Herfarth, H.; Schölmerich, J.; Falk, W.; et al. 13-Oxo-ODE is an endogenous ligand for PPARgamma in human colonic epithelial cells. Biochem. Pharmacol. 2007, 74, 612–622.
- Umeno, A.; Sakashita, M.; Sugino, S.; Murotomi, K.; Okuzawa, T.; Morita, N.; Tomii, K.; Tsuchiya, Y.; Yamasaki, K.; Horie, M.; et al. Comprehensive analysis of PPARγ agonist activities of stereo-, regio-, and enantio-isomers of hydroxyoctadecadienoic acids. Biosci. Rep. 2020, 40, BSR20193767.
- Brunnström, A.; Hamberg, M.; Griffiths, W.J.; Mannervik, B.; Claesson, H.E. Biosynthesis of 14,15-hepoxilins in human l1236 Hodgkin lymphoma cells and eosinophils. Lipids 2011, 46, 69–79.
- Feltenmark, S.; Gautam, N.; Brunnström, A.; Griffiths, W.; Backman, L.; Edenius, C.; Lindbom, L.; Björkholm, M.; Claesson, H.E. Eoxins are proinflammatory arachidonic acid metabolites produced via the 15-lipoxygenase-1 pathway in human eosinophils and mast cells. Proc. Natl. Acad. Sci. USA 2008, 105, 680–685.
- Green, A.R.; Freedman, C.; Tena, J.; Tourdot, B.E.; Liu, B.; Holinstat, M.; Holman, T.R. 5S,15S-Dihydroperoxyeicosatetraenoic Acid (5,15-diHpETE) as a Lipoxin Intermediate: Reactivity and Kinetics with Human Leukocyte 5-Lipoxygenase, Platelet 12-Lipoxygenase, and Reticulocyte 15-Lipoxygenase-1. Biochemistry 2018, 57, 6726–6734.
- Steinhilber, D.; Roth, H.J. New series of lipoxins isolated from human eosinophils. FEBS Lett. 1989, 255, 143–148.
- Tornhamre, S.; Elmqvist, A.; Lindgren, J.A. 15-Lipoxygenation of leukotriene A4. Studies Of 12- and 15-lipoxygenase efficiency to catalyze lipoxin formation. Biochim. Biophys. Acta 2000, 1484, 298–306.
- Fiore, S.; Maddox, J.F.; Perez, H.D.; Serhan, C.N. Identification of a human cDNA encoding a functional high affinity lipoxin A4 receptor. J. Exp. Med. 1994, 180, 253–260.
- Romano, M.; Recchia, I.; Recchiuti, A. Lipoxin receptors. Sci. World J. 2007, 7, 1393–1412.
- Schaldach, C.M.; Riby, J.; Bjeldanes, L.F. Lipoxin A4: A new class of ligand for the Ah receptor. Biochemistry 1999, 38, 7594–7600.
- Russell, R.; Gori, I.; Pellegrini, C.; Kumar, R.; Achtari, C.; Canny, G.O. Lipoxin A4 is a novel estrogen receptor modulator. FASEB J. 2011, 25, 4326–4337.
- Ishizaki, Y.; Morita, I.; Murota, S. Arachidonic acid metabolism in cultured astrocytes from rat embryo and in C6 glioma cells. Brain Res. 1989, 494, 138–142.
- Boado, R.J.; Pardridge, W.M.; Vinters, H.V.; Black, K.L. Differential expression of arachidonate 5-lipoxygenase transcripts in human brain tumors: Evidence for the expression of a multitranscript family. Proc. Natl. Acad. Sci. USA 1992, 89, 9044–9048.
- Golubic, M.; Prayson, R.A.; Vargo, L.; Bondar, J.; Barnett, G.H. Increased expression of 5-lipoxygenase in glioblastoma multiforme. Adv. Exp. Med. Biol. 2003, 525, 205–208.
- Nathoo, N.; Prayson, R.A.; Bondar, J.; Vargo, L.; Arrigain, S.; Mascha, E.J.; Suh, J.H.; Barnett, G.H.; Golubic, M. Increased expression of 5-lipoxygenase in high-grade astrocytomas. Neurosurgery 2006, 58, 347–354.
- Wang, B.; Yu, S.C.; Jiang, J.Y.; Porter, G.W.; Zhao, L.T.; Wang, Z.; Tan, H.; Cui, Y.H.; Qian, C.; Ping, Y.F.; et al. An inhibitor of arachidonate 5-lipoxygenase, Nordy, induces differentiation and inhibits self-renewal of glioma stem-like cells. Stem. Cell Rev. Rep. 2011, 7, 458–470.
- Simmet, T.; Luck, W.; Winking, M.; Delank, W.K.; Peskar, B.A. Identification and characterization of cysteinyl-leukotriene formation in tissue slices from human intracranial tumors: Evidence for their biosynthesis under in vivo conditions. J. Neurochem. 1990, 54, 2091–2099.
- Black, K.L.; Hoff, J.T.; McGillicuddy, J.E.; Gebarski, S.S. Increased leukotriene C4 and vasogenic edema surrounding brain tumors in humans. Ann. Neurol. 1986, 19, 592–595.
- Wang, X.; Chen, Y.; Zhang, S.; Zhang, L.; Liu, X.; Zhang, L.; Li, X.; Chen, D. Co-expression of COX-2 and 5-LO in primary glioblastoma is associated with poor prognosis. J. Neurooncol. 2015, 125, 277–285.
- Jin, T.B.; Li, X.L.; Yang, H.; Jiri, M.; Shi, X.G.; Yuan, D.Y.; Kang, L.L.; Li, S.Q. Association of polymorphisms in FLT3, EGFR, ALOX5, and NEIL3 with glioblastoma in the Han Chinese population. Med. Oncol. 2013, 30, 718.
- Kim, J.A.; Chung, Y.J.; Lee, Y.S. Intracellular Ca2+ mediates lipoxygenase-induced proliferation of U-373 MG human astrocytoma cells. Arch. Pharm. Res. 1998, 21, 664–670.
- Lim, J.Y.; Oh, J.H.; Jung, J.R.; Kim, S.M.; Ryu, C.H.; Kim, H.T.; Jeun, S.S. MK886-induced apoptosis depends on the 5-LO expression level in human malignant glioma cells. J. Neurooncol. 2010, 97, 339–346.
- Souza, F.D.C.; Ferreira, M.T.; Colquhoun, A. Influence of Lipoxygenase Inhibition on Glioblastoma Cell Biology. Int. J. Mol. Sci. 2020, 21, 8395.
- Xu, J.P.; Liu, H.; Bian, X.W.; Chen, J.H.; Zhou, X.D.; Wu, Y.Z. Effect of nordy on biological behaviors of malignant glioma cell line U87MG and the analysis of differential expression proteome. Zhonghua Bing Li Xue Za Zhi 2007, 36, 609–613.
- Ishii, K.; Zaitsu, M.; Yonemitsu, N.; Kan, Y.; Hamasaki, Y.; Matsuo, M. 5-lipoxygenase pathway promotes cell proliferation in human glioma cell lines. Clin. Neuropathol. 2009, 28, 445–452.
- Gáti, I.; Bergström, M.; Csóka, K.; Muhr, C.; Carlsson, J. Effects of the 5-lipoxygenase inhibitors AA-863 and U-60,257 on human glioma cell lines. Prostaglandins Leukot. Essent. Fat. Acids 1990, 40, 117–124.
- Piromkraipak, P.; Parakaw, T.; Phuagkhaopong, S.; Srihirun, S.; Chongthammakun, S.; Chaithirayanon, K.; Vivithanaporn, P. Cysteinyl leukotriene receptor antagonists induce apoptosis and inhibit proliferation of human glioblastoma cells by downregulating B-cell lymphoma 2 and inducing cell cycle arrest. Can. J. Physiol. Pharmacol. 2018, 96, 798–806.
- Abbott, N.J. Blood-brain barrier structure and function and the challenges for CNS drug delivery. J. Inherit. Metab. Dis. 2013, 36, 437–449.
- Chio, C.C.; Baba, T.; Black, K.L. Selective blood-tumor barrier disruption by leukotrienes. J. Neurosurg. 1992, 77, 407–410.
- Black, K.L.; King, W.A.; Ikezaki, K. Selective opening of the blood-tumor barrier by intracarotid infusion of leukotriene C4. J. Neurosurg. 1990, 72, 912–916.
- Black, P.; Hand, C.M.; Vender, J.R.; Finkelstein, S.D. Chemotherapy in experimental brain tumor, part 2: Pretreatment with leukotriene C4 prolongs survival. J. Neurooncol. 1998, 36, 7–19.
- Fellner, S.; Bauer, B.; Miller, D.S.; Schaffrik, M.; Fankhänel, M.; Spruss, T.; Bernhardt, G.; Graeff, C.; Färber, L.; Gschaidmeier, H.; et al. Transport of paclitaxel (Taxol) across the blood-brain barrier in vitro and in vivo. J. Clin. Investig. 2002, 110, 1309–1318.
- Doan, P.; Nguyen, P.; Murugesan, A.; Subramanian, K.; Konda Mani, S.; Kalimuthu, V.; Abraham, B.G.; Stringer, B.W.; Balamuthu, K.; Yli-Harja, O.; et al. Targeting Orphan G Protein-Coupled Receptor 17 with T0 Ligand Impairs Glioblastoma Growth. Cancers 2021, 13, 3773.
- Nguyen, P.; Doan, P.; Rimpilainen, T.; Konda Mani, S.; Murugesan, A.; Yli-Harja, O.; Candeias, N.R.; Kandhavelu, M. Synthesis and Preclinical Validation of Novel Indole Derivatives as a GPR17 Agonist for Glioblastoma Treatment. J. Med. Chem. 2021, 64, 10908–10918.
- Hu, Y.; Luo, H.; Zhu, X.; Guo, H. CRNDE/ETS1/GPR17 Facilitates the Proliferation, Migration, and Invasion of Glioma. Comput. Math. Methods Med. 2021, 2021, 7566365.
- Terpinskaya, T.I.; Osipov, A.V.; Kryukova, E.V.; Kudryavtsev, D.S.; Kopylova, N.V.; Yanchanka, T.L.; Palukoshka, A.F.; Gondarenko, E.A.; Zhmak, M.N.; Tsetlin, V.I.; et al. α-Conotoxins and α-Cobratoxin Promote, while Lipoxygenase and Cyclooxygenase Inhibitors Suppress the Proliferation of Glioma C6 Cells. Mar. Drugs 2021, 19, 118.
- Ding, X.Z.; Tong, W.G.; Adrian, T.E. 12-lipoxygenase metabolite 12(S)-HETE stimulates human pancreatic cancer cell proliferation via protein tyrosine phosphorylation and ERK activation. Int. J. Cancer 2001, 94, 630–636.
- Guo, A.M.; Liu, X.; Al-Wahab, Z.; Maddippati, K.R.; Ali-Fehmi, R.; Scicli, A.G.; Munkarah, A.R. Role of 12-lipoxygenase in regulation of ovarian cancer cell proliferation and survival. Cancer Chemother. Pharmacol. 2011, 68, 1273–1283.
- Zohn, I.E.; Klinger, M.; Karp, X.; Kirk, H.; Symons, M.; Chrzanowska-Wodnicka, M.; Der, C.J.; Kay, R.J. G2A is an oncogenic G protein-coupled receptor. Oncogene 2000, 19, 3866–3877.
- Higuchi, Y.; Yoshimoto, T. Arachidonic acid converts the glutathione depletion-induced apoptosis to necrosis by promoting lipid peroxidation and reducing caspase-3 activity in rat glioma cells. Arch. Biochem. Biophys. 2002, 400, 133–140.
- Strakova, N.; Ehrmann, J.; Dzubak, P.; Bouchal, J.; Kolar, Z. The synthetic ligand of peroxisome proliferator-activated receptor-gamma ciglitazone affects human glioblastoma cell lines. J. Pharmacol. Exp. Ther. 2004, 309, 1239–1247.
- Grommes, C.; Landreth, G.E.; Sastre, M.; Beck, M.; Feinstein, D.L.; Jacobs, A.H.; Schlegel, U.; Heneka, M.T. Inhibition of in vivo glioma growth and invasion by peroxisome proliferator-activated receptor gamma agonist treatment. Mol. Pharmacol. 2006, 70, 1524–1533.
- Liu, Y.; Shi, L.; Liu, Y.; Li, P.; Jiang, G.; Gao, X.; Zhang, Y.; Jiang, C.; Zhu, W.; Han, H.; et al. Activation of PPARγ mediates icaritin-induced cell cycle arrest and apoptosis in glioblastoma multiforme. Biomed. Pharmacother. 2018, 100, 358–366.
- Viita, H.; Pacholska, A.; Ahmad, F.; Tietäväinen, J.; Naarala, J.; Hyvärinen, A.; Wirth, T.; Ylä-Herttuala, S. 15-Lipoxygenase-1 induces lipid peroxidation and apoptosis, and improves survival in rat malignant glioma. In Vivo 2012, 26, 1–8.
- Yuan, H.; Li, M.Y.; Ma, L.T.; Hsin, M.K.; Mok, T.S.; Underwood, M.J.; Chen, G.G. 15-Lipoxygenases and its metabolites 15(S)-HETE and 13(S)-HODE in the development of non-small cell lung cancer. Thorax 2010, 65, 321–326.
- Liu, L.; Li, X.; Shi, J.; Li, L.; Wang, J.; Luo, Z.Z. Effects of FPR2 gene silencing on the proliferation, migration and invasion of human glioma U87 cells. Zhonghua Zhong Liu Za Zhi 2018, 40, 659–666.
- He, H.Q.; Ye, R.D. The Formyl Peptide Receptors: Diversity of Ligands and Mechanism for Recognition. Molecules 2017, 22, 455.
More
