Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Hong-gyu Kang and Version 2 by Catherine Yang.
Plant basic helix-loop-helix (bHLH) transcription factors are involved in many physiological processes, and they play important roles in the abiotic stress responses. bHLH transcription factors are among the superfamilies that are commonly found in plants and animals. The conserved bHLH domain contains approximately 60 amino acids (aa), including a basic DNA binding region and two amphipathic α-helices that are separated by a loop region with a variable length. The basic region consists of the first 15 amino acids. Most bHLH proteins have a glutamic acid residue at position 9 (E9), which can interact with the CA nucleotides in the DNA sequence.
- bHLH transcription factor
- plant growth and development
- plant metabolism synthesis
1. bHLH Transcription Factors Are Responsible for Plant Growth and Development
bHLH transcription factors regulate plant growth and development. Researchers have demonstrated bHLH gene functions in Arabidopsis, as summarized by Hao et al. [1][65]. However, few bHLH genes have been characterized in other crop plants. Here, thwe researchers focus on the functionally characterized bHLH genes in different plants, and mainly in crops (Figure 12).
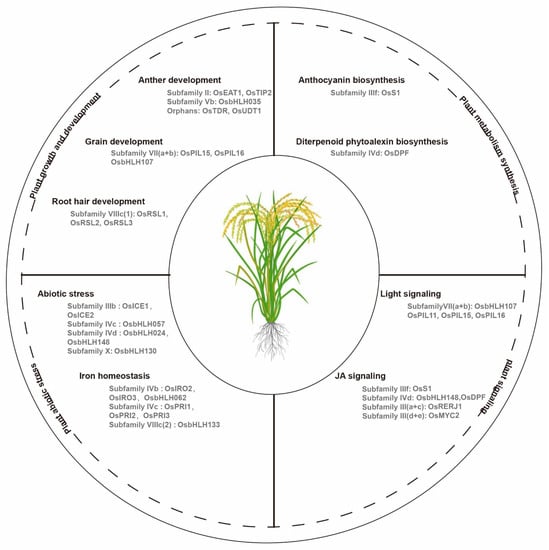
Figure 12. Functional classification of basic helix-loop-helix (bHLH) transcription factors in rice. bHLH genes are involved in various physiological processes for growth, metabolism, signaling, and adaptation to environmental stress.
The members of the same subfamily always have similar functions. In flowering plants, the anther is a reproductive organ that is composed of meiocytes and four cell layers: the epidermis, endothecium, middle layer, and tapetum [2][66]. The rice bHLH transcription factor in subfamily II, TDR Interacting Protein 2 (TIP2), plays an important role in the formation of the middle layer and tapetum during early anther development through the regulation of Tapetum Degeneration Retardation (TDR) and Eternal Tapetum 1 (EAT1). TDR and EAT1 are the key regulators in rice tapetal programmed cell death [3][67]. EAT1, which is a conserved bHLH transcription factor in land plants, is involved in this complex process via the direct activation of the transcriptions of two aspartic protease encoding genes (AP25 and AP37) [4][68]. In addition, EAT1 also regulate meiotic phasiRNA biogenesis in anther tapetum, and Undeveloped Tapetum 1(UDT1) is a potential interacting partner of both EAT1 and TIP2 during the early meiosis process in rice [5][6][69,70]. Furthermore, rice TIP2 and EAT1 belong to subfamily II, which indicate that the bHLH genes in subfamily II may act as the key transcriptional regulators in anther development. The overexpression of OsbHLH35 endows plants with small and curved anthers, which result in a reduction in the seed production (Figure 12) [7][71]. In this process, three Growth Regulating Factor (GRF) family members, including OsGRF3, OsGRF4, and OsGRF11, act as the transcriptional regulators of OsbHLH35, and OsGRF11 acts as a negative regulator of OsbHLH35 in rice. In other plants, LoUDT1 from the oriental lily hybrid Siberia (Lilium spp.), which is the homologous gene of OsUDT1, is also related to anther development [8][72]. Citrullus lanatus Abnormal Tapetum 1 (ClATM1), which is the first male sterility gene in watermelon, encodes a bHLH protein, and plays important role in the regulation of anther development, which researchers verified via CRISPR/Cas9-mediated mutagenesis [9][73]. In tomato, Solyc01g081100, which is a homolog of OsEAT1, is the candidate gene for the dysfunctional pollen and tapetum development in the male sterile 32 (ms32) mutant, and the CRISPR/Cas9-mediated modification of the bHLH protein encoded gene Solyc02g079810 causes male sterility in tomato plants [10][11][74,75].
The antagonist of the PGL1 (APG) of rice in subfamily VII(a+b), called OsPIL16, controls the grain length and weight [12][76]. OsPIL16 interacts with two atypical bHLH transcription factors, Regulator of Grain Length 1 (PGL1) and PGL2, to antagonistically regulate the development of the rice grain length [12][13][76,77]. The overexpression of PGL1 in lemma/palea increased the grain length and weight in transgenic rice [13][77]. In addition, OsPIL15 shares a close genetic relationship with OsPIL16, which also influences cell division by affecting the transport of cytokinin (CTK), which results in decreased cell numbers in rice grains [14][78].
Root hairs are long tubular projections of trichoblasts, which are the hair-forming cells on the epidermis of the plant root. They increase the plant surface area to improve absorption of nutrients from soil [15][16][81,82]. The rhizoid root hairs are essential for ion exchange, anchorage functions, and microbial interactions in the soil of land plants [15][81]. Various bHLH transcription factors participate in this critical root development process [17][83]. During root hair cell differentiation, the hair and non-hair cells are differentiated from morphologically identical epidermal cells [18][84]. The root hair cells produce an outside long tubule from the hair-forming cells on the epidermis of the plant root, and they function in the absorption of nutrients and water, and in interaction with microbes [16][17][82,83]. In
Arabidopsis root hair defective 6 (Atrhd6) and root hair defective six-like1 (Atrsl1) double mutants that lack the RSL class I gene function, transformation with 35S: OsRSL1, 35S:OsRSL2 or 35S:OsRSL3 restored the expressions of the RSL class II genes, which indicates the functional conservation of the RSL genes between rice and Arabidopsis in root hair development [19][85]. In Brachypodium distachyon, the RSL class I genes, including BdRSL1, BdRSL2, and BdRSL3 also promote root hair development [20][86]. In sweet sorghum, researchers identified a new atypical bHLH transcription factor, SbbHLH85, in subfamily VIIIc(2), as a key gene for root development via increase in the numbers and lengths of the root hair via ABA and auxin signaling pathways [21][87].
2. bHLH Transcription Factors Play Important Roles in Plant Metabolism Synthesis
Anthocyanins are the major pigments of flavonoid compounds, and they endow plants with colors, such as blue, purple, and red, in many flowers, fruits, and vegetables [22][23][88,89]. In plants, anthocyanins attract pollinators or seed dispersers, and they protect against UV radiation, pathogen attacks, and abiotic stresses [24][25][90,91]. Furthermore, anthocyanins are compounds with potential health-benefits for lowering the risk of cardiovascular diseases, certain cancers, and diabetes in humans due to high levels of antioxidant activity [26][27][28][92,93,94]. The MBW complex (R2R3-MYB, bHLH, and WDR) mediates the anthocyanin biosynthetic pathway, which is one of the most conserved and well-studied secondary metabolism pathways in plants [29][30][95,96].
The functionally regulated bHLH genes in anthocyanin biosynthesis are primarily classified as subfamily IIIf. Petroni and Tonelli [24][90] and Jaakola [31][97] reviewed the bHLH genes that are involved in anthocyanin biosynthesis in various horticultural species, such as petunia, antirrhinum, and grape. In rice, Sun et al. [32][98] explored the minimal MBW members required for anthocyanin biosynthesis, including S1 (bHLH), C1 (MYB), and WA1 (WD40). Under chromium stress, the rice MBW complex is regulated by the jasmonate (JA) signal and represses anthocyanin accumulation in tissues (Figure 12) [33][99]. Additionally, low temperature induced SlAH in subfamily IVd regulates anthocyanin biosynthesis in tomatoes to protect young seedlings from cold stress, which indicates the bHLH transcription factor functional connections between anthocyanin biosynthesis and abiotic stress [34][100]. In Freesia hybrida, subfamily IIIf, the bHLH transcription factors FhTT8L and FhGL3L interact with FhMYB5 in proanthocyanidin biosynthesis during flower pigmentation [35][101]. Peach PpbHLH3 regulated the anthocyanin biosynthesis with MYB10.1 and MYB10.3 in fruit development during ripening (at the transition from the S3 to S4 stage) [36][102].
In addition, the rice bHLH transcription factor Diterpenoid Phytoalexin Factor (DPF) in subfamily IVd positively regulates the expressions of the diterpenoid phytoalexin (DP) biosynthesis genes in the process of DP accumulation. DPF was the first bHLH transcription factor to be characterized in DP biosynthesis through the N-box (5′-CACGAG-3′) [37][104]. In an orphan group from tomato, the bHLH transcription factor SlAR plays an important role in the carotenoid biosynthesis in the fruits, which may particularly affect lycopene accumulation [38][105].
3. bHLH Transcription Factors Are Involved in Plant Signaling
Green plants obtain most of their energy from light through photosynthesis, and light is an important environmental factor that determines plant growth and development. Plants can sense the red, far-infrared, and blue light spectra through photoreceptor systems, including phytochromes (PHYs), cryptochromes (CRYs), and phototropins (PHOTs), and they can then mediate the transcriptional networks in the light-regulated processes [39][40][41][42][43][106,107,108,109,110]. In Arabidopsis, members of the phytochrome-interacting factors (PIFs) in subfamily VII(a+b), such as PIF1/ PIF-like (PIL5), PIF3, PIF4, PIF5/PIL6, PIF6/PIL2, and PIF7, can interact with PHYs and play central roles in light signaling regulation [1][65].
Rice has six PILs (OsPIL11 to OsPIL16) in subfamily VII(a+b), and some of the members are involved in the light signaling pathways [42][43][44][45][109,110,111,112]. OsPIL15 was negatively regulated by light with the onset of light exposure in etiolated seedlings [46][113]. The overexpression of OsPIL15 produced exhibited shorter above-ground parts, an undeveloped root system, smaller tiller angles, and enhanced shoot gravitropism, which were related to the skotomorphogenesis development, which was likely regulated by the auxin [47][48][114,115]. In addition, Li et al. [49][116] demonstrated that the fusion of the SRDX transcriptional repressor motif in the C-terminal of OsPIL11 and OsPIL16 caused constitutively photomorphogenic phenotypes with short coleoptiles and open leaf blades in darkness. Nevertheless, OsPIL16 was able to bind to the N-box region of the OsDREB1B promoter, and it was substantially induced by cold stress in a phyB mutant [50][117]. In peach, PpPIF8 can interact with PpDELLA2 through an unknown motif, and the overexpression of PpPIF8 in Arabidopsis promoted increases in the plant height and branch numbers [51][118].
Phytohormones, such as JA and abscisic acid (ABA), regulate plant growth, development, and defense processes. JA triggers the degradation of the protein jasmonate ZIM-domain (JAZ) by 26S protease, which induces the activation of multiple downstream JA-mediated responsive genes [52][53][54][55][119,120,121,122]. Some bHLH transcription factors are involved in the hormone signaling pathways in plants. RERJ1, which is a rice JA-responsive gene in subfamily III(a+c), was up-regulated via exposure to wounding or drought stress [56][123]. Interestingly, RERJ1 also interacted with OsMYC2 to mediate the defense processes against herbivory and bacterial infection through JA signaling [57][124]. The DPF in rice was also induced by JA, and as well as response to blast fungus infection, copper chloride, and UV light [37][104]. Additionally, JA-regulated OsMYC2 induced the expressions of insect defense-related genes, and it simultaneously activated some of the biosynthetic pathways for the defense-related metabolites in rice, which was proven in a knockdown osmyc2RNAi plant (Figure 12) [57][124]. In Artemisia annua, AaMYC2-Like, the methyl jasmonate (MeJA) responsive transcription factor, played a prominent role in regulating the artemisinin biosynthetic pathway, as researchers confirmed through the transient overexpression of AaMYC2-Like in the leaves [58][125]. Furthermore, two MYC-type bHLH transcription factors from A. annua, AabHLH2 and AabHLH3 act as transcription repressors and functional redundantly to regulate artemisinin biosynthesis [59][126]. ABA plays a central role in a variety of physiological processes, and it is also a key abiotic stress-related hormone that responds to various environmental stresses in plants, such as cold, drought, and salt. Several bHLH transcription factors in subfamily IIIb are involved in abiotic stresses via the ABA signaling pathway. A group of PebHLH genes in moso bamboo (Phyllostachys edulis) possess various cis-elements for ABA and JA in their promoters, which are up-regulated by biotic and abiotic stresses, ABA and MeJA stimuli [60][31].
Under specific conditions, multiple phytohormones and environmental factors are constantly cross talking to affect plant growth and development. Based on previous studies, light usually promotes cell expansion in plant growth, while ABA and JA are normally involved in the biotic and abiotic stress responses. Some bHLH transcription factors play important roles in signal transduction networks that are mediated by plant hormones. Phytohormones and environment-responsive bHLH transcription factors participate in various plant developmental processes by interacting with each other either cooperatively or antagonistically to modulate plant growth.
4. bHLH Transcription Factors Are Related to Plant Abiotic Stress and Iron Homeostasis
Plants have evolved to adapt to the stressful conditions that are unfavorable for their growth and development [61][62][63][64][127,128,129,130]. However, these adverse environmental conditions substantially affect the yields of crops such as rice [65][66][67][131,132,133]. Cold stress impacts plant productivity, especially during the flowering stage, which substantially lowers the probability of reproductive success. Cold stress signals can be perceived by putative sensors and induce the expressions of stress-responsive genes to modulate the cellular activities through the cytosolic Ca2+ levels [67][133]. bHLH genes that are related to plant abiotic stresses are mainly in subfamily IIIb, including OsICE1 and OsICE2, which researchers have widely identified in recent years.
In rice, OsbHLH148 responds to the initial JA signal and regulates drought responsive genes, including OsDREBs and OsJAZs, endowing the rice with drought tolerance [68][134]. BEAR1, which is a bHLH gene, regulates the salt response, which was demonstrated by the salt-sensitive phenotypes of its knockdown or knockout transgenic rice [69][135]. The osbhlh024 mutant (A91), with a nucleotide base deletion generated by the CRISPR/Cas9 strategy, increased the shoot weight, and produced high antioxidant activities under salt stress, which indicates that OsbHLH024 might play a negative role in the salt stress response of rice [70][136]. In addition, OsWIH2, which is a drought induced WIH gene, was activated by OsbHLH130, and the overexpression of OsbHLH130 resulted in substantially higher drought tolerance via its participation in cuticular wax biosynthesis, with reductions in the water loss rate and ROS accumulation (Figure 12) [71][137]. OsbHLH057 targeted the AATCA cis-element, in the promoter of Os2H16, a gene responding to fungal attack in rice, and overexpression of OsbHLH057 enhanced rice disease resistance to fungus Rhizoctonia solani and drought tolerance [72][138]. In sorghum, a typical bHLH gene, SbbHLH85, plays a key role in the root development, and its overexpression increased Na+ absorption, which indicates that SbbHLH85 might play a negative regulatory role in salt tolerance [21][87].
The ectopic expression of CabHLH035 in pepper (Capsicum annuum L.) enhanced the salt tolerance in transgenic Arabidopsis [73][139]. The overexpression of MxbHLH18 in apple increased iron and high-salinity stress tolerances in Arabidopsis [74][140]. Zuo et al. [75][34] reported the genome-wide identification of the bHLH family genes in Zoysia japonica. The expressions of ZjbHLH62/ZjICE2, ZjbHLH67, ZjbHLH76/ZjICE1, ZjbHLH88, ZjbHLH97, and ZjbHLH120 in subfamily IIIb were affected by cold, salt, dehydration, and/or ABA [75][34]. Furthermore, the overexpression of ZjICE1 and ZjICE2 endowed transgenic Arabidopsis and Z. japonica plants with abiotic stress tolerance via the activation of the DREB/CBF regulon, and they also enhanced ROS scavenging [76][77][141,142]. The overexpression of MfbHLH38, which is a bHLH gene of Myrothamnus flabellifolia, increased the tolerance to drought and salt stresses in transgenic Arabidopsis [78][143]. HbICE2, which is a novel ICE-like gene in the rubber tree (Hevea brasiliensis), is involved in JA-mediated cold tolerance and its overexpression enhanced the cold resistance in transgenic Arabidopsis [79][144]. The ThbHLH1 (subfamily XI) plays an important role in stress signaling pathways and induces the expressions of stress-related genes [80][145]. The bHLH transcription factors regulate a wide range of plant growth and stress response signaling pathways, and some of them share homeopathic pathways for plant survival under unfavorable or stressful conditions.
Metal deficiency (iron) substantially affects many physiological processes of plants, such as photosynthesis and respiration. Both low and high concentrations of iron greatly affect the growth of plants. Plants have developed regulatory systems to control their iron uptake and maintain their Fe homeostasis, and bHLH transcription factors play important roles in this process [81][146]. In rice, the Iron-related transcription factor 3 (OsIRO3) in subfamily IVb is upregulated by environmental stress, and OsIRO3 is characterized as a negative regulator. The overexpression of OsIRO3 reduced the gene expression in the process of Fe chelator biosynthesis that included OsIRO2 [82][147]. OsbHLH133, in the subfamily VIIIc(2), plays an important role in the Fe-deficiency signaling network under Fe-deficient conditions in rice [83][151]. ThWe researchers present the results for other species, such as soybean, and apple. Manganese (Mn) is also an essential micro-nutrient that acts as a cofactor in the redox reactions in photosynthesis. ZmbHLH105 confers improved Mn tolerance on the transgenic tobacco by repressing the expressions of the Mn/Fe-regulated transporter genes to reduce the Mn acclamation [84][152].
In summary, plant growth is strictly controlled by intricate regulation mechanisms, and the bHLH family genes always function with many other proteins to allow plants to perform specific developmental processes at suitable times, and to increase their chances of survival under unfavorable environments. Various research studies have demonstrated that bHLH transcription factors play important roles in a broad range of plant growth and developmental processes via crosstalk. For example, RERJ1 mediates the defense processes against herbivory and bacterial infection through JA signaling [57][124], OsPIL11 and OsPIL16 are involved in photomorphogenic developmental processes that are affected by light [49][116], and OsbHLH148 regulates drought stress in rice, which also responds to JA treatment [68][134]. The establishment of this mechanism is complicated, and systematic investigations into the bHLH genes in plants will facilitate their use for crop improvement programs.
