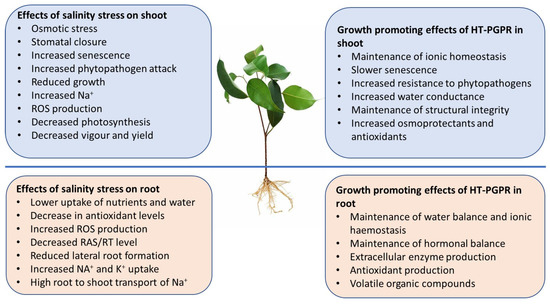
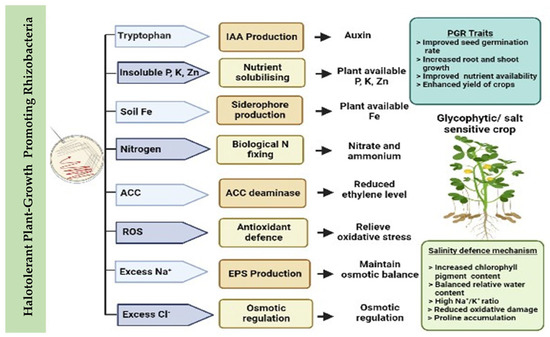
Soil salinity is one of the major abiotic constraints in agricultural ecosystems worldwide. High salinity levels have negative impacts on plant growth and yield, and affect soil physicochemical properties. Salinity also has adverse effects on the distribution and abundance of soil microorganisms. Halotolerant plant growth-promoting rhizobacteria (HT-PGPR) secrete secondary metabolites, including osmoprotectants, exopolysaccharides, and volatile organic compounds. The importance of these compounds in promoting plant growth and reducing adverse effects under salinity stress has been widely recognised. HT-PGPR are emerging as effective biological strategies for mitigating the harmful effects of high salinity; improving plant growth, development, and yield; and remediating degraded saline soils.
- exopolysaccharides
- osmoprotectants
- growth hormones
- soil microbes
- volatile organic compounds
1. Introduction
2. HT-PGPR: Diversity and Their Effect on Crop Production
3. HT-PGPR and Their Effects in Mitigating Salt Stress in Crops
HT-PGPR provide plants with resistance to salt stress through several key processes. One such process is the regulation of the salt overly sensitive (SOS) pathway, which is involved in salt influx/efflux across membranes, through metabolites and associated gene expression patterns. It has been shown that the SOS1 gene is directly regulated by metabolites such as EPS, VOCs, and suitable solutes (i.e., proline, glycine betaines, and trehalose) [25], which also direct stress regulation in SOS genes [3], HKT1 transporter (high-affinity K) expression [32], and other genes implicated in the reduction in salt stress, i.e., ethylene biosynthesis and antioxidant protein encoding genes [33][34][33,34]. Examples of the beneficial effects of HT-PGPR, along with how they help plants cope with saline conditions in different plants, are reviewed and presented in Table 1.| Plant Species |
HT-PGPR Species |
Gene/s Involved |
Mechanism to Mitigate Stress | Effect Observed | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana L. | Bacillus oryzicola YC7007 | RD22, KIN1, RD29B, RD20, RD22, and ERD1 | Stem and the root of the seedlings released stress-related genes | Enhanced plant tolerance to salt stress | [35] |
| Pseudomonas putida PS01 |
APX2 and GLYI7 | APX2 and GLYI7 genes were downregulated | ABA signalling, jasmonic acid production route, ROS scavenging, detoxification | [36] | |
| Pseudomonas knackmussii MLR6 | NHX1, HKT1, SOS2, SOS3, SAG13, and PR1 | Enhanced stomatal conductance, transpiration rate, chlorophyll, and carotenoid levels | Reduced electrolyte leakage and priming ROS accumulation increasing cell membrane stability | [37] | |
| Bacillus amyloliquefaciens SQR9 | NHX1 and NHX7 | Involved in reducing GSH biosynthesis | Reduced ion toxicity by sequestering Na+ into vacuoles and releasing Na+ from the cell | [38] | |
| Burkholderia phytofirmans PsJN | Upregulation of RD29A and GLYI7, and downregulation of LOX2 | Enhancement of proline and transcription of genes related to abscisic acid signalling and downregulated gene Lipoxygenase 2 | Abscisic acid signalling, ROS reduction, detoxifying, jasmonate synthesis, and ion transport | [39] | |
| Paenibacillus yonginensis DCY84T | AtRSA1, AtVQ9 and AtWRKY8 | Upregulated salt-stress genes | Promoted more resistance to salinity, drought, and aluminium stresses | [40][41][40,41] | |
| Enterobacter sp. EJ01 | DREB2b, RD29A, RD29B, RAB18, P5CS1, P5CS2, MPK3, and MPK6 | Upregulated salt-stress genes | Promoted more resistance to salinity and enhanced plant growth | [42] | |
| Bacillus subtilis GB03 | HKT1 | Down- and upregulates HKT1 in roots and shoots, respectively | Decreased total plant Na+ accumulation | [23] | |
| Bacopa monneri L. | Dietzia natronolimnaea STR1 | SOS1, SOS4, TaST, TaNHX1, TaHAK, and TaHKT1 | Reduction in ABA-signalling, upregulated TaABARE and TaOPR1 | Abscisic acid signalling, ROS scavenging, antioxidant enzyme activity, enhanced ion transporter expression, high K+/Na+ ratio | [24] |
| Bacillus pumilus STR2, Exiguobacterium oxidotolerens STR36 |
- | Mixture of plant growth-promoting traits under primary and secondary saline condition | Produced higher yield, high proline/lipid content peroxidation | [2] | |
| Cicer arietinum L. | Planococcus rifietoensis (RT4) and Halomonas variabilis (HT1) | - | Biofilm and exopolysaccharides production | Improved crop growth, soil aggregation, and soil fertility | [43] |
| Glycine max L. | Arthrobacter woluwensis AK1 | - | Reduced endogenous ABA and controlled antioxidant activity | Mitigated salinity stress and increased plant growth | [44] |
| Microbacterium oxydans, Arthrobacter woluwensis, Arthrobacter aurescens, Bacillus aryabhattai, and Bacillus megaterium |
- | Increased production of IAA, GA, siderophores, and phosphate solubilisation | Increased antioxidant enzymes and K absorption; reduced Na+ in plant tissue; phytohormone | [45] | |
| Pseudomonas simiae AU | P5CS, PPO and HKT1 | Downregulated HKT1, LOX, PPO, and P5CS genes | Increased chlorophyll, phosphate solubilisation, IAA, and siderophores; decreased root surface in saline | [46] | |
| Pseudomonas sp. strain AK-1 | HTK1 | Improve K+/Na+ ratio and Exopolysaccharide production binds free Na+ from soil | Increased shoot/root length and decreased Na+/K+ ratio | [33] | |
| Pseudomonas simiae AU | VSP2 | Increase vegetative storage protein (VSP), gamma-glutamyl hydrolase (GGH), and RuBisCo proteins | Reduced Na, increased K and P in soybean seedling roots, high proline and chlorophyll content | [47] | |
| Helianthus annuus L. | Pseudomonas libanensis TR1 | - | ACC-deaminase and exopolysaccharide production | Ni and Na+ accumulation potential increased along with plant growth. | [48] |
| Pseudomonas spp. | - | Upregulating of ACC deaminase | Improved P and K contents, and K+/Na+ ratio in shoot | [49] | |
| Hordeum vulgare L. | Bacillus mojavensis, B. pumilus and Pseudomonas fluorescens | S1 and S3 | ACC deaminase, IAA, and proline production | Reduced plant Na concentration, stimulated root growth, improved water and nutrient absorption | [50] |
| B. aryabhattai MS3 | BZ8, SOS1, GIG, and NHX1 | Increased salt stress resistance and accumulation | Adaptation of plant under saline condition | [51] | |
| Bacillus amyloliquefaciens SN13 | DHN | Upregulated salt stress-responsive genes and protein-related genes | Lipid peroxidation and electrolyte leakage reduced; increased rice biomass, water content, proline, and total soluble sugar | [52] | |
| Bacillus megaterium ST2-1 | - | IAA production | Stimulated the growth of rice roots and dry biomass | [53] | |
| Pseudomonas pseudoalcaligenes ST1, Bacillus pumilus ST2 |
EU440977 and FJ840535 | Accumulation of proline decrease with inoculation, antioxidative activity | Enhanced plant growth by ROS scavenging and higher accumulation of osmoprotectant | [54] | |
| Puccinellia tenuiflora L. | Bacillus subtilis (GB03) |
- | Upregulated PtHKT1;5 and PtSOS1 genes, downregulated PtHKT2;1 |
Na homeostasis modulation, exclusive K+ absorption | [55] |
| Solanum lycopersicum L. | Leclercia adecarboxylata MO1 | - | ACC deaminase and IAA production | Increased soluble sugars: organic glucose, sucrose, fructose, malic, amino acid, and proline | [56] |
| Sphingobacterium sp. BHU-AV3 | - | Reduction in ROS concentration in plant | Enhanced antioxidant activities and energy metabolism | [57] | |
| Enterobacter sp. EJ01 | DREB2b, RD29A, RD29B, and RAB18 | Downregulated P5CS1 and P5CS2, and upregulated MPK3 and MPK6 | Biosynthesis, defence pathway modulation, salt response |
[42] | |
| Pseudomonas putida UW4 | Toc GTPase | Toc GTPase genes were upregulated and reduction in ACC deaminase | Increased shoot length and chlorophyll concentration | [58] | |
| Trifolium repens L. | Bacillus subtilis (GB03) | - | Reduced shoot and root Na+, improving K+/Na+ ratio | Decreased Na+, increased chlorophyll, leaf osmotic potential, cell membrane integrity | [59] |
| Triticum aestivum L. | Pseudomonas aeruginosa GI-1, and Burkholderia gladioli GI-6 | - | P solubilisation, catalase activity, IAA production, N assimilation, and siderophores production | Encouraged growth and yield and improve soil fertility | [60][61][60,61] |
| Arthrobacter nitroguajacolicus | - | Upregulated 152 genes whereas 5 genes were downregulated | Amplified ACC, IAA, siderophore, and phosphate solubility. ROS detoxification, Na+ homeostasis, abiotic stress | [62] | |
| Serratia marcescens CDP-13 | - | Increased salt tolerance in plant | ACC deaminase, phosphate solubilisation, siderophore, indole acetic acid, N fixation, and ammonia synthesis | [63] | |
| Pseodomonas sp and Enterobacter cloacae (R-10) | B-22 and S-49 | K and Zn solubilisation for identifying antifungal activity | Enhanced K+ uptake, dry matter of wheat | [64] | |
| Hallobacillus sp. SL3 Bacillus halodenitrificans PU62 |
acdS | IAA production and siderophore production, phosphate solubilising, and siderophore production | Increased root elongation and dry weight | [65] | |
| Zea mays L. | Serratia liquefaciens KM4 | Upregulation of stress-related genes (APX, CAT, SOD, RBCS, RBCL, H+-PPase, HKT1, and NHX1) | Regulating redox potential and stress-related gene expression | Higher leaf gas exchange, osmoregulation, antioxidative defence mechanisms, and nutrient uptake boosted maize growth and biomass production | [66] |
| Azospirillum lipoferum, Azospillum sp., Azotobacter chroococcum, Azotobacter sp., and Bacillus sp. | - | Exopolysaccharide inoculation in the soil | Increased root and shoot dry weights, chlorophyll and carotenoids, restricted Na and Cl uptake, and increased shoot N, P, and K | [67] | |
| Abelmoschus esculentus L. |
Enterobacter sp. UPMR18 |
X55749 | ROS pathway upgradation and enhancement in antioxidant enzyme activities | Higher germination, growth, and chlorophyll improved salt tolerance | [68] |
