Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Camila Xu and Version 1 by Melissa Marie Rowe.
Non-coding RNAs (ncRNAs) encompass all RNAs that are not translated into proteins. They are classified by features such as length and structure, and include microRNAs (miRs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs).
- non-coding RNA
- microRNA
- long non-coding RNA
1. Introduction
One topic of particular interest in human health is epigenetic regulation by non-coding RNAs. Non-coding RNAs (ncRNAs) encompass all RNAs that are not translated into proteins. They are classified by features such as length and structure, and include microRNAs (miRs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). These different classes of ncRNAs participate in complex interaction networks with each other, mRNAs, and various signaling pathways to enact changes on cells, surrounding tissue, and whole organ systems.
2. The Major Types of Non-Coding RNAs
MicroRNAs (miRs) are the smallest of the non-coding RNAs, with an average size of 22 nucleotides [11][1]. A primary miR (pri-miR), which may be kilobases long, is transcribed from its genomic coding region [12][2]. The Microprocessor complex trims and processes the pri-miR to create a 60–90 nucleotide precursor miR (pre-miR) [12][2]. The pre-miR often exists as a double-stranded hairpin loop and is exported into the cytoplasm by Exportin 5 bound to Ran-GTP (see Figure 1A) [13][3]. Two mature miRs are created when the Dicer nuclease splits and processes the strands [13][3]. These mature miRs are then referred to as the “5p” or “3p” species, coming from the 5′ and 3′ ends of the pre-miR transcript, respectively [14][4]. However, not all mature miRs are created equal. Often, either the 5p or 3p strand is expressed more highly and has a stronger effect on epigenetic gene regulation (and is thus referred to as the “guide strand”), while the other “passenger strand” is frequently (but not always) degraded [13][3]. Advancing technology has recently established that stably expressed passenger strands are more common than previously thought, and further studies are warranted to uncover unknown functions of these miR species [14][4].
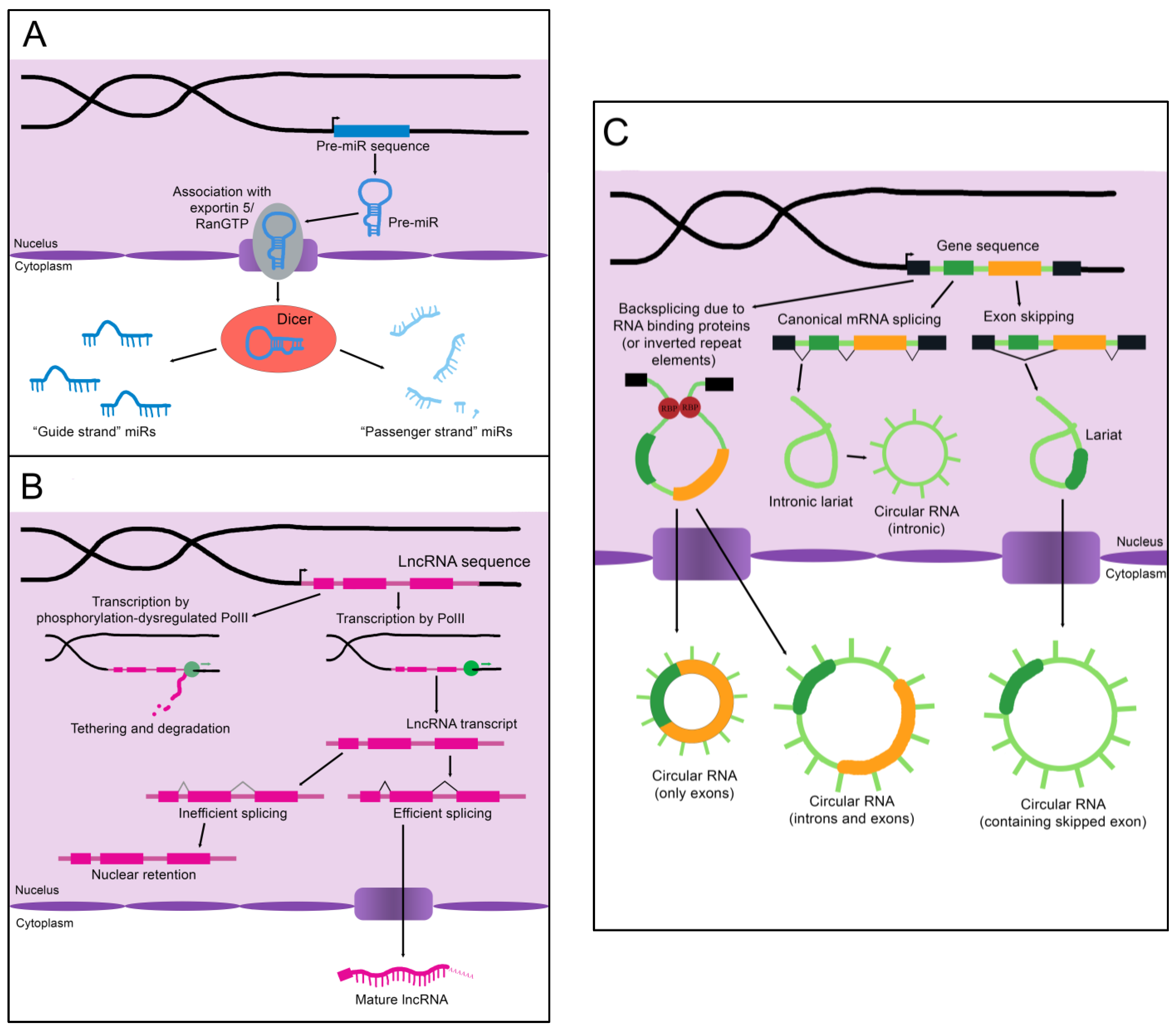
Figure 1. The biogenesis of non-coding RNAs. (A) An immature miR transcript is transcribed and exported into the cytoplasm by Exportin 5 bound to Ran-GTP. The “guide” and “passenger” strands are created when the Dicer nuclease splits and processes these precursors. (B) The sequence of a lncRNA is transcribed by RNA polymerase II (PolII) and either spliced to its mature form or inefficiently spliced and retained in the nucleus. Alternatively, the sequence is transcribed by phosphorylation-dysregulated PolII, remains tethered to the chromatin, and either accumulates over time or is degraded. (C) CircRNAs are created through back-splicing mediated by RNA binding proteins, inverted repeat elements, or lariats created during mRNA splicing. The mature circRNA may contain only exons, only introns, or a combination of both. RBP: RNA-binding protein.
Following processing, the mature miR associates with Argonaute protein (Ago), transactivation response element RNA-binding protein (TRBP), protein kinase RNA activator (PACT), and the still-bound Dicer to create the microRNA-induced silencing complex (miRISC), which carries out the epigenetic suppression of target mRNAs (Figure 2A) [13][3]. The identification of target mRNAs is not trivial and currently cannot be done reliably using only computational tools. This is because only 6–8 consecutive nucleotides in the miR’s 5′ “core” sequence are required to bind the microRNA recognition element (MRE) in a target mRNA [13][3]. In addition, imperfect base pairing can occur between the core sequence and MREs [13][3]. MiR to MRE interactions are also able to make G:U pairs, providing additional flexibility in target binding [13][3]. Due to the variable nature of this process, a given miR can have hundreds of potential mRNA targets [13][3]. Fortunately, biochemical methods such as CLIP-Seq (cross-linking immunoprecipitation followed by high throughput sequencing) have been developed that greatly facilitate the identification of miR-mRNA targeting relationships, and these have been applied to the study of liver regeneration [15][5]. Depending on the miR/mRNA pair, miRs can either cause degradation of the mRNA or lead to the inhibition of its translation [11][1].
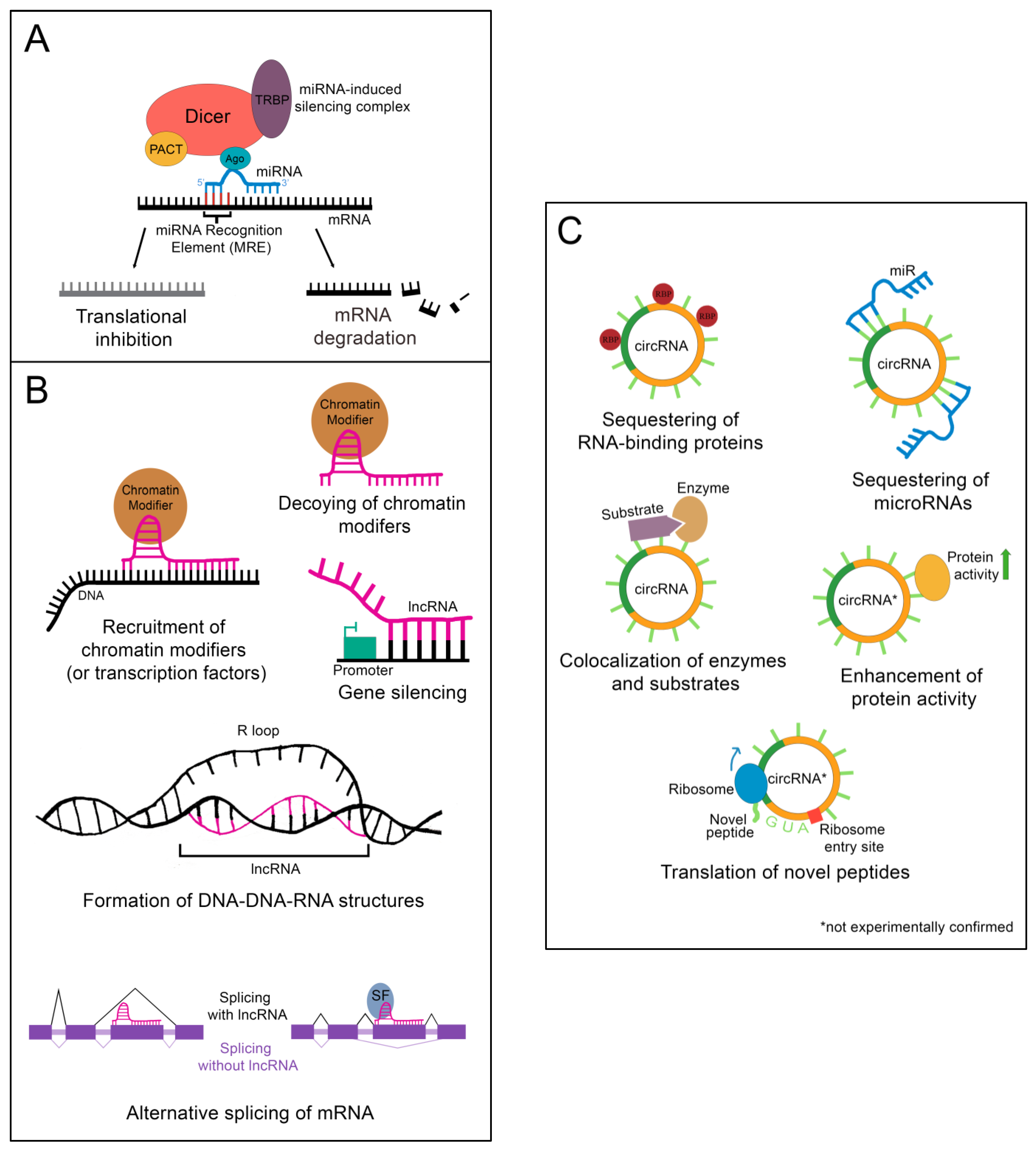
Figure 2. Epigenetic regulation by ncRNAs. (A) The microRNA-induced silencing complex downregulates mRNAs through translational inhibition or degradation. TRBP: transactivation response element RNA-binding protein; PACT: protein kinase RNA activator; Ago: Argonaute (B) lncRNAs alter gene expression by binding DNA, binding mRNA, or by sequestering chromatin modifiers. DNA binding by lncRNAs can result in chromatin modifier or transcription factor recruitment, gene silencing, or the formation of DNA-DNA-RNA structures. Binding of a lncRNA to mRNA often leads to alternative splicing of the mRNA. For instance, the presence of a lncRNA on an exon can lead to that exon being skipped. Alternatively, recruitment of splicing factors by a bound lncRNA can lead to the retention of an exon that would otherwise be skipped. SF: splicing factor (C) CircRNAs function as sponges for RNA-binding proteins or microRNAs. Some can also colocalize enzymes with their substrates. A few are proposed to enhance protein activity or be translated into novel peptides, although these functions have yet to be experimentally validated. RBP: RNA-binding protein.
As the name implies, long non-coding RNAs (lncRNAs) are more sizable than miRs, operationally defined as being at least 200 nucleotides in length [16][6]. Some estimates for the number of lncRNAs encoded in humans exceed 100,000, although the number that are actually transcribed or functional is unknown [16][6]. LncRNAs resemble protein-coding mRNAs in both their length and nuclear processing (many possess a 5′ cap and 3′ polyadenylation and have introns); however, they are processed less efficiently [16][6]. LncRNAs are more likely than mRNAs to be transcribed by phosphorylation-dysregulated Pol II (Figure 1B). This can lead to lncRNA transcripts becoming tethered to chromatin, where they are either degraded or accumulate over time [16][6]. The splicing signals of lncRNAs are also weaker, and therefore lncRNAs are frequently retained in the nucleus due to inefficient splicing [16][6].
Importantly, nuclear localization does not mean that a given lncRNA is biologically inert. LncRNAs impact gene regulation through a wide array of physical interactions (Figure 2B). LncRNA-chromatin interactions are capable of recruiting or decoying chromatin modifiers, and of interacting directly with the DNA to form DNA-DNA-RNA triplexes and R-loops, both of which have unique effects on gene regulation [16][6]. LncRNAs are also well known to silence gene expression. This can serve to balance gene dosage, most famously in the silencing of the second mammalian female X chromosome through lncRNA XIST in a process termed “X-inactivation” [16,17][6][7]. LncRNAs can also act on gene expression through alterations to gene accessibility or recruitment of transcription factors [16][6]. Finally, lncRNAs have been shown to alter pre-mRNA splicing through direct association with mRNAs or recruitment or decoying of splicing factors [16,18][6][8].
Circular RNAs (circRNAs), self-joined RNA molecules, have proven difficult to isolate and characterize due to their lack of the canonical features of mature RNA, such as a polyadenylated 3′ end [19][9]. However, with continuing advances in sequencing technology, the role of these unique molecules in disease pathology and homeostasis is beginning to be appreciated. There are multiple ways circRNAs can arise in a cell (Figure 1C). Inverted repeat elements or RNA-binding proteins that flank one or more exons can lead to back-splicing. This results in a circular RNA molecule containing either solely exons or both exons and introns [19][9]. Back-splicing can also occur in lariats created during exon-skipping, leading to a circRNA containing the skipped exon(s) [19][9]. Lastly, lariats that escape debranching in canonical linear splicing can self-join to become circRNAs containing only introns [19][9]. The majority of exonic circRNAs are exported to the cytoplasm, while circRNAs containing introns tend to remain in the nucleus [19][9]. Since these molecules do not have exposed ends like most RNA molecules, they are extremely stable and tend to accumulate with age [19][9].
Although most circRNAs remain uncharacterized, those that have been studied in-depth have revealed multiple potential mechanisms of action (Figure 2C). The majority of those analyzed thus far have been proposed to function as sponges for miRs, reducing their effective concentration [19][9]. CircRNAs are also able to decoy RNA-binding proteins, and a few have been shown to bind enzymes and their substrates, promoting colocalization and activity [19][9]. Although not yet experimentally confirmed, circRNAs may also functionally enhance or recruit proteins [19][9]. Some circRNAs contain internal ribosome entry sites and AUG codons, making it possible to be translated, which could give rise to unique peptides that would not be possible through canonical RNA processing [19][9].
In addition to biological functions in their cell of origin, ncRNAs are sometimes secreted from the cell, typically in exosomes. The protection of the exosomal membrane allows ncRNAs to remain relatively stable in the extracellular space, and exosomal ncRNAs have been detected in both the blood and urine of human patients [20][10]. Given this stability, and the ability of cells to uptake exosomes produced by distal tissues, it has been proposed that exosomal ncRNAs are able to exert biological effects far from their cell of origin [21,22][11][12]. The ability to monitor ncRNA levels through the non-invasive collection of body fluids has made exosomal ncRNAs prime candidates to act as biomarkers for various disease states.
Numerous studies have found changes in non-coding RNAs in various liver pathologies, several of which are outlined in Table 1. Emphasis has been placed on RNA species that are highly expressed in the liver, whether at baseline or in proliferative states, or that have experimentally proven impacts on liver cell biology reflected in multiple sources. Examined pathologies and injury states mainly affect hepatocyte function.
Table 1.
Summary of major ncRNA-associated liver pathologies.
| ncRNA | Disease Association(s) | Other Notable Function(s) | Citations |
|---|---|---|---|
| miR-122 | Hepatitis C | [23,24,25,26,27,28][13][14][15][16][17][18] | |
| NASH | |||
| miR-21 | NASH | Limits regenerative ability | [29,[30,31,1932,33]][20][21][22][23] |
| Hepatocellular carcinoma | |||
| let-7a | Hepatocellular carcinoma | Altered following partial hepatectomy | [34,35,36][24][25][26] |
| Cholangiocarcinoma | |||
| let-7b | Hepatitis C | Downregulated in small volume liver grafts | [36,37,38,39][26][27][28][29] |
| Acetaminophen overdose | Altered following partial hepatectomy | ||
| Hepatocellular carcinoma | |||
| let-7c | Acetaminophen overdose | Altered following partial hepatectomy | [36,37][26][27] |
| let-7d | Acetaminophen overdose | Altered following partial hepatectomy | [36,37][26][27] |
| let-7e | Altered following partial hepatectomy | [36][26] | |
| let-7f | Downregulated in small volume liver grafts | [36][26] | |
| Altered following partial hepatectomy | |||
| let-7g | Hepatitis C | Altered following partial hepatectomy | [36,40,41,42,43][26][30][31][32][33] |
| Hepatocellular carcinoma | |||
| let-7i | Altered following partial hepatectomy | [36][26] | |
| miR-451a | Chronic hepatitis B | [44,45][34][35] | |
| Hepatocellular carcinoma | |||
| miR-144 | Chronic hepatitis B | [44,45][34][35] | |
| Hepatocellular carcinoma | |||
| ZSCAN16-AS1 (lncRNA) | Hepatocellular carcinoma | [46][36] | |
| MALAT1 (lncRNA) | Hepatocellular carcinoma NAFLD/NASH Fibrosis |
Liver regeneration | [47,3748][][38] |
References
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Conrad, T.; Ntini, E.; Lang, B.; Cozzuto, L.; Andersen, J.B.; Marquardt, J.U.; Ponomarenko, J.; Tartaglia, G.G.; Vang Orom, U.A. Determination of primary microRNA processing in clinical samples by targeted pri-miR-sequencing. RNA 2020, 26, 1726–1730.
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell. Pharmacol. 2011, 3, 83–92.
- Choo, K.B.; Soon, Y.L.; Nguyen, P.N.; Hiew, M.S.; Huang, C.J. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 2014, 21, 95.
- Schug, J.; McKenna, L.B.; Walton, G.; Hand, N.; Mukherjee, S.; Essuman, K.; Shi, Z.; Gao, Y.; Markley, K.; Nakagawa, M.; et al. Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. BMC Genom. 2013, 14, 264.
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118.
- Sahakyan, A.; Yang, Y.; Plath, K. The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol. 2018, 28, 999–1013.
- Pisignano, G.; Ladomery, M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. Noncoding RNA 2021, 7, 21.
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691.
- Xu, Y.X.; Pu, S.D.; Li, X.; Yu, Z.W.; Zhang, Y.T.; Tong, X.W.; Shan, Y.Y.; Gao, X.Y. Exosomal ncRNAs: Novel therapeutic target and biomarker for diabetic complications. Pharmacol. Res. 2022, 178, 106135.
- Isaac, R.; Reis, F.C.G.; Ying, W.; Olefsky, J.M. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021, 33, 1744–1762.
- Wang, D.; Zhang, W.; Zhang, C.; Wang, L.; Chen, H.; Xu, J. Exosomal non-coding RNAs have a significant effect on tumor metastasis. Mol. Ther. Nucleic Acids 2022, 29, 16–35.
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005, 309, 1577–1581.
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schuttler, C.G.; Fehr, C.; Junemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008, 27, 3300–3310.
- Cheung, O.; Puri, P.; Eicken, C.; Contos, M.J.; Mirshahi, F.; Maher, J.W.; Kellum, J.M.; Min, H.; Luketic, V.A.; Sanyal, A.J. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology 2008, 48, 1810–1820.
- Long, J.K.; Dai, W.; Zheng, Y.W.; Zhao, S.P. miR-122 promotes hepatic lipogenesis via inhibiting the LKB1/AMPK pathway by targeting Sirt1 in non-alcoholic fatty liver disease. Mol. Med. 2019, 25, 26.
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98.
- Huang, R.; Duan, X.; Fan, J.; Li, G.; Wang, B. Role of Noncoding RNA in Development of Nonalcoholic Fatty Liver Disease. Biomed. Res. Int. 2019, 2019, 8690592.
- van Raalte, D.H.; Li, M.; Pritchard, P.H.; Wasan, K.M. Peroxisome proliferator-activated receptor (PPAR)-alpha: A pharmacological target with a promising future. Pharm. Res. 2004, 21, 1531–1538.
- Loyer, X.; Paradis, V.; Henique, C.; Vion, A.C.; Colnot, N.; Guerin, C.L.; Devue, C.; On, S.; Scetbun, J.; Romain, M.; et al. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARalpha expression. Gut 2016, 65, 1882–1894.
- Juskeviciute, E.; Dippold, R.P.; Antony, A.N.; Swarup, A.; Vadigepalli, R.; Hoek, J.B. Inhibition of miR-21 rescues liver regeneration after partial hepatectomy in ethanol-fed rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 311, G794–G806.
- Ng, R.; Song, G.; Roll, G.R.; Frandsen, N.M.; Willenbring, H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J. Clin. Investig. 2012, 122, 1097–1108.
- Correia de Sousa, M.; Calo, N.; Sobolewski, C.; Gjorgjieva, M.; Clement, S.; Maeder, C.; Dolicka, D.; Fournier, M.; Vinet, L.; Montet, X.; et al. Mir-21 Suppression Promotes Mouse Hepatocarcinogenesis. Cancers 2021, 13, 4983.
- Liu, L.; Zhao, J.; Peng, Y.; Yang, M.; Zhang, L.; Jin, X. miR-let-7a-5p Inhibits Invasion and Migration of Hepatoma Cells by Regulating BZW2 Expression. Onco Targets Ther. 2020, 13, 12269–12279.
- Pan, X.; Wang, G.; Wang, B. MicroRNA-1182 and let-7a exert synergistic inhibition on invasion, migration and autophagy of cholangiocarcinoma cells through down-regulation of NUAK1. Cancer Cell Int. 2021, 21, 161.
- Chen, X.; Murad, M.; Cui, Y.Y.; Yao, L.J.; Venugopal, S.K.; Dawson, K.; Wu, J. miRNA regulation of liver growth after 50% partial hepatectomy and small size grafts in rats. Transplantation 2011, 91, 293–299.
- Wang, K.; Zhang, S.; Marzolf, B.; Troisch, P.; Brightman, A.; Hu, Z.; Hood, L.E.; Galas, D.J. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. USA 2009, 106, 4402–4407.
- Hui, L.; Zheng, F.; Bo, Y.; Sen-Lin, M.; Ai-Jun, L.; Wei-Ping, Z.; Yong-Jie, Z.; Lei, Y. MicroRNA let-7b inhibits cell proliferation via upregulation of p21 in hepatocellular carcinoma. Cell Biosci. 2020, 10, 83.
- Cheng, J.C.; Yeh, Y.J.; Tseng, C.P.; Hsu, S.D.; Chang, Y.L.; Sakamoto, N.; Huang, H.D. Let-7b is a novel regulator of hepatitis C virus replication. Cell. Mol. Life Sci. 2012, 69, 2621–2633.
- Chen, K.J.; Hou, Y.; Wang, K.; Li, J.; Xia, Y.; Yang, X.Y.; Lv, G.; Xing, X.L.; Shen, F. Reexpression of Let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. Biomed. Res. Int. 2014, 2014, 742417.
- Ji, J.; Zhao, L.; Budhu, A.; Forgues, M.; Jia, H.L.; Qin, L.X.; Ye, Q.H.; Yu, J.; Shi, X.; Tang, Z.Y.; et al. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J. Hepatol. 2010, 52, 690–697.
- Lan, F.F.; Wang, H.; Chen, Y.C.; Chan, C.Y.; Ng, S.S.; Li, K.; Xie, D.; He, M.L.; Lin, M.C.; Kung, H.F. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int. J. Cancer 2011, 128, 319–331.
- Chou, W.W.; Huang, C.F.; Yeh, M.L.; Tsai, Y.S.; Hsieh, M.Y.; Huang, C.I.; Huang, J.F.; Tsai, P.C.; Hsi, E.; Juo, S.H.; et al. MicroRNA let-7g cooperates with interferon/ribavirin to repress hepatitis C virus replication. J. Mol. Med. 2016, 94, 311–320.
- Loukachov, V.V.; van Dort, K.A.; Maurer, I.; Takkenberg, R.B.; de Niet, A.; Reesink, H.W.; Willemse, S.B.; Kootstra, N.A. Identification of Liver and Plasma microRNAs in Chronic Hepatitis B Virus infection. Front. Cell. Infect. Microbiol. 2022, 12, 790964.
- Zhao, J.; Li, H.; Zhao, S.; Wang, E.; Zhu, J.; Feng, D.; Zhu, Y.; Dou, W.; Fan, Q.; Hu, J.; et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol. Cancer 2021, 20, 46.
- Lv, C.; Wan, Q.; Shen, C.; Wu, H.; Zhou, B.; Wang, W. Long noncoding RNA ZSCAN16AS1 promotes the malignant properties of hepatocellular carcinoma by decoying microRNA451a and consequently increasing ATF2 expression. Mol. Med. Rep. 2021, 24, 780.
- Lu, J.; Guo, J.; Liu, J.; Mao, X.; Xu, K. Long Non-coding RNA MALAT1: A Key Player in Liver Diseases. Front. Med. 2021, 8, 734643.
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640.
More
