Gastric cancers have been historically classified based on histomorphologic features. The Cancer Genome Atlas network reported the comprehensive identification of genetic alterations associated with gastric cancer, identifying four distinct subtypes— Epstein-Barr virus (EBV)-positive, microsatellite-unstable/instability (MSI), genomically stable and chromosomal instability. In particular, EBV-positive and MSI gastric cancers seem responsive to novel immunotherapies drugs. The aim of this entreviewy is to describe MSI and EBV positive gastric cancer’s subgroups and their relationship with novel immunotherapy.
- MSI
- EBV
- molecular subtypes
- immunotherapy
1. Molecular Classification
Recent progress in genomic technology has now allowed GCs to be studied at the molecular level facilitating the identification of potentially “druggable” alterations in GC, such as gene mutations, chromosomal alterations, transcriptional changes and epigenetic derangements (Table 1).
Table 1. Molecular classification of Gastric Cancer according TCGA (The Cancer Genome Atlas)emt; ACRG (Asian Cancer Research Group) and Li et al. Classification.
|
|
SUBTYPES |
MOLECULAR FEATURES |
|
|
|
|
|
TCGA (The Cancer Genome Atlas) |
EBV (9%) |
-DNA hypermethylation, including CDKN2A (p16) but not MLH1 promoters - PIK3CA mutations - JAK2 gene amplification -PDL1/PDL2 overexpression |
|
|
MSI (22%) |
-high mutation rate -DNA methylation with epigenetic silencing of MLH1 - Hypermutation of many genes including HLA class 1 factors |
|
|
GS (20%) |
-molecular alterations in cell adhesion/ cell migration pathways -ARID1 and BCOR mutations |
|
|
CIN (50%) |
- chromosomal instability (CIN) -amplification of genes (most encoding tyrosine kinase receptors) |
|
ACRG (Asian Cancer Research Group) |
MSS/TP53 + (26%) |
-frequent EBV positivity -intermediate mutation rate |
|
|
MSI (23%) |
-high mutation rate |
|
|
EMT (15%) |
-low mutation rate -loss of epithelial markers |
|
|
MSS/TP53- (36%) |
-TP53 mutations -genomic instability |
|
Li et al. Classification |
REGULAR C1 |
-2.4 mutations /megabase; range, 0–8.3 - TP53, XIRP2, APC mutations
|
|
|
REGULAR C2 |
-2.4 mutations /megabase; range, 0–8.3 - ARID1A, CDH1, PIK3CA, ERBB2, RHOA mutations - |
|
|
HYPERMUTATED |
-20.5 mutations/megabase; range, 9.6–200.2) |
|
|
SUBTYPES |
MOLECULAR FEATURES |
|
|
|
|
|
TCGA (The Cancer Genome Atlas) |
EBV (9%) |
-DNA hypermethylation, including CDKN2A (p16) but not MLH1 promoters - PIK3CA mutations - JAK2 gene amplification -PDL1/PDL2 overexpression |
|
|
MSI (22%) |
-high mutation rate -DNA methylation with epigenetic silencing of MLH1 - Hypermutation of many genes including HLA class 1 factors |
|
|
GS (20%) |
-molecular alterations in cell adhesion/ cell migration pathways -ARID1 and BCOR mutations |
|
|
CIN (50%) |
- chromosomal instability (CIN) -amplification of genes (most encoding tyrosine kinase receptors) |
|
ACRG (Asian Cancer Research Group) |
MSS/TP53 + (26%) |
-frequent EBV positivity -intermediate mutation rate |
|
|
MSI (23%) |
-high mutation rate |
|
|
EMT (15%) |
-low mutation rate -loss of epithelial markers |
|
|
MSS/TP53- (36%) |
-TP53 mutations -genomic instability |
|
Li et al. Classification |
REGULAR C1 |
-2.4 mutations /megabase; range, 0–8.3 - TP53, XIRP2, APC mutations
|
|
|
REGULAR C2 |
-2.4 mutations /megabase; range, 0–8.3 - ARID1A, CDH1, PIK3CA, ERBB2, RHOA mutations - |
|
|
HYPERMUTATED |
-20.5 mutations/megabase; range, 9.6–200.2) |
2. MSI and EBV Positive Gastric Cancer’s Subgroups
In 2014, a milestone research carried out by The Cancer Genome Atlas (TCGA) network reported the comprehensive identification of genetic alterations associated with GC, testing 295 frozen GC tissues from untreated patients with six different molecular platforms, including whole exome sequencing (WES), messenger RNA sequencing, microRNA sequencing (miRNA), array-based DNA methylation profiling, reverse-phase protein array and array-based somatic copy number analysis. On the basis of an integrative analysis of this molecular information, the TCGA team identified four distinct subtypes—EBV-positive (8.8%), which displayed recurrent PIK3CA mutations, extreme DNA hypermethylation and amplification of JAK2, CD274 (also known as PD-L1) and PDCD1LG2 (also known as PD-L2), microsatellite-unstable/instability (MSI, 21.7%), which shows elevated mutation rates, including mutations of genes encoding targetable oncogenic signalling proteins, genomically stable (19.7%), which were enriched for the diffuse histological variant and mutations of RHOA, fusions involving RHO-family GTPase-activating proteins, CDH1 somatic mutations, CLDN18–ARHGAP6 or ARHGAP26 fusions and chromosomal instability (CIN, 49.8%), which showed marked aneuploidy and focal amplification of receptor tyrosine kinases. Interestingly, neither racial nor survival differences were found among each subgroup[1].
In the subsequent year, the Asian Cancer Research Group (ACRG) proposed a new molecular classification for GC[2]. On the basis of whole-genome sequencing, gene expression profiling, genome-wide copy number microarrays and targeted gene sequencing, the ACRG first divided the GCs into MSI and microsatellite stable (MSS) types. Then, secondarily, the MSS GC were divided into epithelial-mesenchymal transition (EMT), TP53+ and TP53- groups.
MSS/EMT was characterized by the worst prognosis and a higher chance of recurrence (63%) which occurs mainly in peritoneum; it was predominantly associated with Lauren diffuse histologic type (> 80%), diagnosed at a younger age and clinical stage III/IV. ACRG MSS/EMT subtype had a partial overlap with GS subtype in TCGA. On the contrary, MSI subtype occurred predominantly at the antrum (75%) and over 60% were diagnosed as the intestinal subtype and at an early stage (I/II). Moreover, MSI subgroup showed the best prognosis, followed by MSS/TP53+ and MSS/TP53-. EBV infection occurred more frequently in the MSS/TP53- group than in other groups.
A comparison between TCGA and ACRG data allows distinguishing both similarities and differences. Both TCGA and ACRG classifications identify MSI as a separate subgroup of GC. GS was associated with MSS/EMT, EBV to MSS/TP53+ and CIN to MSS/TP53- but it was unlikely to find a complete correspondence among these other subgroups. While CIN and GS TCGA subtypes tumors were present across all ACRG subtypes, TCGA GS, EBV+ and CIN subtypes were enriched in ACRG MSS/EMT, MSS/TP53+ and TP53− subtypes, respectively.
According to histological features, the majority of TCGA diffuse subtype cases (57%) belonged to GS subgroup but only 27% of the cases were present in the correspondent ACRG MSS/EMT subgroup, suggesting that TCGA diffuse subtype case were less heterogeneous. In TCGA, EBV positive GCs represented a distinctive subgroup, whereas in ACRG cohort EBV infection was found more frequently in MSS/TP53+ subtype, without hypermethylation or hypermutation. PI3K mutations and ARID1A mutations were less prevalent in MSS than in EBV positive subtype.
As previously reported, these two classifications had a partial overlap. Several differences, including geografic differences in the two patient populations (Eastern vs Western patients) and differences in tumor sampling, amplified by the use of distinct platforms, have been considered responsible for the mismatch across categories of these two classifications.[3][4]In a more recent paper, Li et al. proposed a novel classification system, based on the aggregation of somatic molecular profiles of 544 GC patients from previous genomic studies. GCs were divided into regular (86.8%; 2.4 mutations/Mb; range, 0–8.3) and hypermutated (13.2%; 20.5 mutations/Mb; range, 9.6–200.2) subtypes based on mutation burden, the latter of which showed a marked overrepresentation of samples with microsatellite instability. The regular type was subclassified into 2 subgroups, C1 and C2, with distinct clinical outcomes, independently of disease staging. C1 carried mutations in TP53, XIRP2 and APC and correlated with significantly better prognosis, while C2 overexpressed mutations in ARID1A, CDH1, PIK3CA, ERBB2 and RHOA[5]. Interestingly, in this analysis, CDH1 mutations were found as an independent prognostic factor in diffuse-type but not intestinal-type GC regardless of TNM staging. Unfortunately, the application in the clinical setting of these molecular classifications is still limited. The highly complex methodology, lack of prospective validation and short period of follow-up limit the reproducibility in standard laboratories lacking cutting edge technologies. Nevertheless, the great amount of data obtained from these multi-omics profiling classifications has helped to redefine the genetic and pathogenic landscape of GC and to elucidate novel molecular targets and therapeutic strategies paving the way for precision oncology. Moreover, these classifications are expected to improved patient stratification for novel clinical trials. The molecular classification of GCs will probably change the way of thinking in GC oncology. Because of the somewhat promising targeted therapies that are under investigation in clinical trials, EBV positive GCs and MSI GCs seem to have the highest importance[6], in particular we will focus on EBV and MSI subgroups that seem to have the most clinically relevant impact (Figure 1).
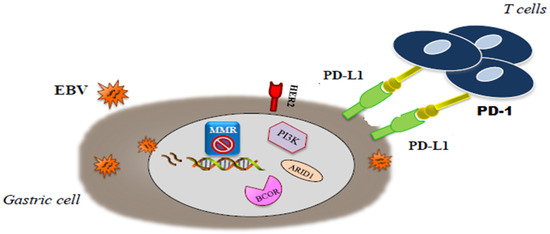
Figure 1. Molecular interaction in microsatellite-unstable/instability (MSI) end . Epstein-Barr virus (EBV) gastric cancer (GC).
References
- The Cancer Genome Atlas Research Network; Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202-209, 10.1038/nature13480.
- Razvan Cristescu; Jeeyun Lee; Michael Nebozhyn; Kyoung-Mee Kim; Jason C Ting; Swee Seong Wong; Jiangang Liu; Yong Gang Yue; Jian Wang; Kun Yu; et al.Xiang S YeIn-Gu DoShawn LiuLara GongJake FuJason Gang JinMin-Gew ChoiTae Sung SohnJoon Ho LeeJae-Moon BaeSeung Tae KimSe Hoon ParkInSuk SohnSin-Ho JungPatrick TanRonghua ChenJames HardwickWon Ki KangMark AyersDai HongyueChristoph ReinhardAndrey LobodaSung KimAmit Aggarwal Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nature Medicine 2015, 21, 449-456, 10.1038/nm.3850.
- Lionel Kankeu Fonkoua; Nelson S. Yee; Molecular Characterization of Gastric Carcinoma: Therapeutic Implications for Biomarkers and Targets. Biomedicines 2018, 6, 32, 10.3390/biomedicines6010032.
- Yoku Hayakawa; Nilay Sethi; Antonia R. Sepulveda; Adam J. Bass; Timothy C. Wang; Oesophageal adenocarcinoma and gastric cancer: should we mind the gap?. Nature Reviews Cancer 2016, 16, 305-318, 10.1038/nrc.2016.24.
- Xiangchun Li; William K.K. Wu; Rui Xing; Sunny Hei Wong; Yuexin Liu; Xiaodong Fang; Y. L. Zhang; Mengyao Wang; Jiaqian Wang; Lin Li; et al.Yong ZhouSenwei TangShaoliang PengKunlong QiuLongyun ChenKexin ChenHuanming YangWei ZhangMatthew T.V. ChanYouyong LuJoseph J. Y. SungJun Yu Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Research 2016, 76, 1724-1732, 10.1158/0008-5472.can-15-2443.
- Yu Sunakawa; Heinz-Josef Lenz; Molecular Classification of Gastric Adenocarcinoma: Translating New Insights from The Cancer Genome Atlas Research Network. Current Treatment Options in Oncology 2015, 16, 331, 10.1007/s11864-015-0331-y.
