The human body harbors trillions of microbes of different kinds performing various physiological activities, such as priming the immune system, influencing host metabolism, and improving health by providing important metabolites such as short-chain fatty acids. Although the gut is considered the “microbial organ” of our body as it hosts the most microbes, there are microbes present in various other important anatomical locations differing in numbers and type. Research has shown the presence of microbes in utero, sparking a debate on the “sterile womb” concept, and there is much scope for more work in this area. It is important to understand the early-life microbiome colonization, which has a role in the developmental origins of health and disease in later life.
- microbiome
- amniotic fluid
- placenta
- genitals
- neonates
- metabolic health
1. Introduction
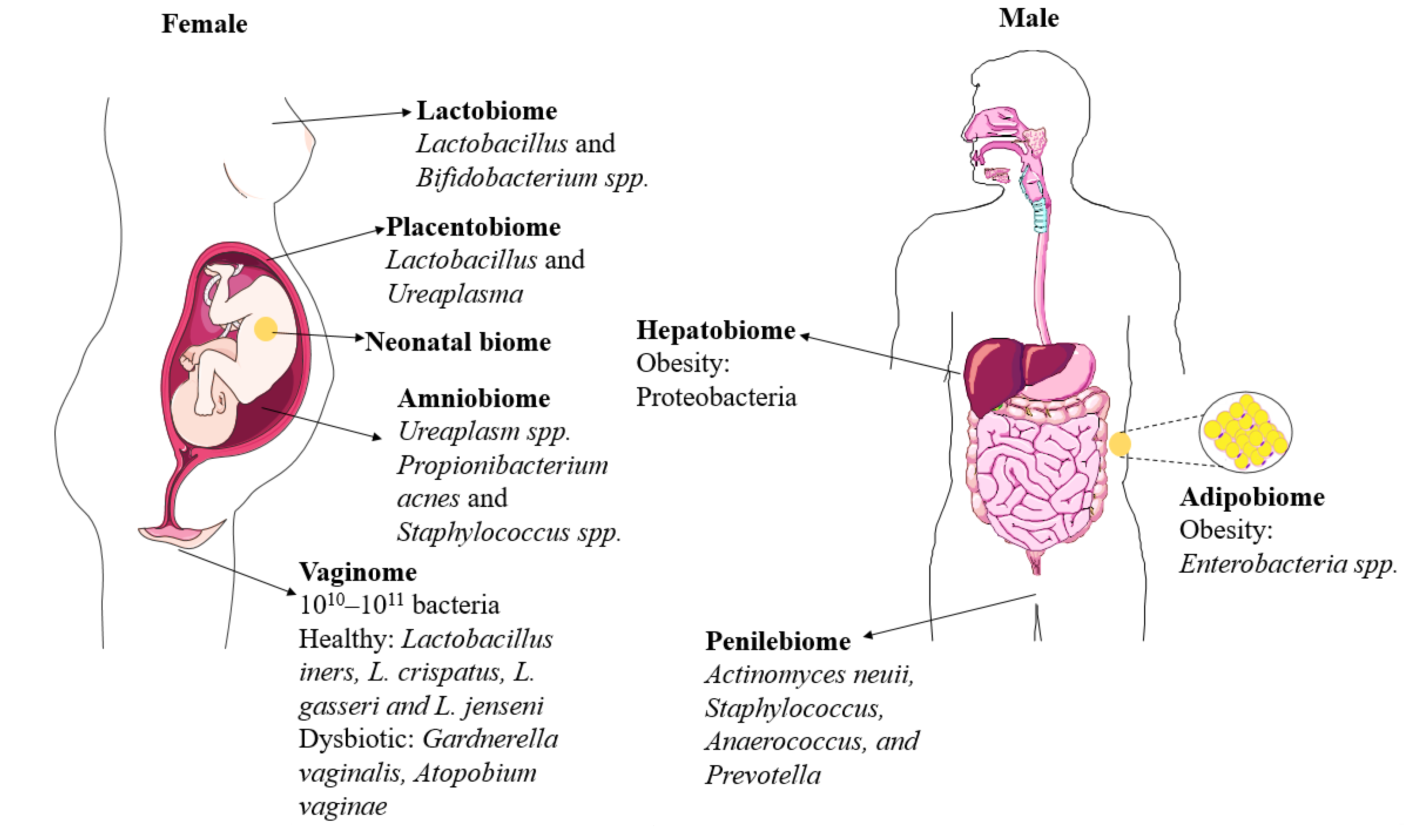
2. Genital Microbiomes
2.1. Vaginobiome
| Biome | Study Type | Dominant Bacteria | Method Used | Reference |
|---|---|---|---|---|
| 1. Vaginome | Human (pregnant and non-pregnant) | Pregnant: Lactobacillus vagitypes (L. crispatus, L. iners, L. gasseri and L. jensenii) Non-pregnant: Lactobacillus |
V1–V3 16S rRNA | [11] |
| Vaginal microbial profiles in European (E) | Pre-pregnancy: L. crispatus Pregnancy: L. jensenii, L. crispatus PP: BV- associated taxa Prevotella spp., Clostridium spp., Atopobium spp. and Megasphaera spp. |
V1-V2 16S rRNA | [12] | |
| Vaginal microbial profiles in African American (AA) versus European (E) ancestry women | BV: Gardnerella vaginalis (AA) AA: L. iners E: L. crispatus, L. iners, G. vaginalis AA/E differences: Mycoplasma, Gardnerella, Prevotella and Sneathia |
V1–V3 16S rRNA | [13] | |
| Vaginome during pregnancy, preterm and PP | Pregnancy: L. crispatus, L. gasseri, L. iners, L. jensenii Preterm: Gardnerella and Ureaplasma PP: Peptoniphilus, Prevotella, and Anaerococcus |
V3–V5 16S rRNA | [15] | |
| 2. Penilebiome | Penile (both meatal and glans/coronal sulcus/circumcised) and vaginal microbial profiles related to BV | Penile: Corynebacterium (circumcised), Streptococcus, Anaerococcus, Finegoldia. BV: Parvimonas, L. iners, L. crispatus, Fastidiosipila, and Prevotella |
V3–V4 16S rRNA | [14] |
| Vaginal and penile microbiomes related to herpes simplex virus type 2 (HSV-2) | BV: G. vaginalis and L. iners Penile: Ureaplasma and Aerococcus (HSV-2) |
V3–V4 16S rRNA | [16] | |
| 3. Amniobiome | Preterm in 2nd trimester, asymptomatic | Ureaplasma and/or Mycoplasma spp. | 16S rRNA | [17] |
| In utero to first 4 days of birth | Enterobacter, Escherichia/Shigella and Propionibacterium | V1-V3 16S rRNA | [5,18][5][18] | |
| 4. Placentalbiome | In utero to first 4 days of birth | Propionibacterium, Enterobacter and Escherichia/Shigella | V1-V3 16S rRNA | [5] |
| Meconium in twins | Salinibacter and Enterobacteriaceae_unclassified | V3-V4 16S rRNA | [19] | |
| 5. Meconiobiome | In utero to first 4 days of birth | Propionibacterium, Escherichia/Shigella, and Lactobacillus | V1-V3 16S rRNA | [5] |
| Meconium in twins | Enterobacteriaceae_unclassified | V3-V4 16S rRNA | [19] | |
| Temporal and spatial variation in early-life microbiome | Lactobacillus, Bifidobacterium, Staphylococcus, and Enterococcus spp. | V3–V5 16S | [15] | |
| 6. Lactobiome | Milk microbiome | Staphylococcus and Streptococcus | V1-V3 16S rRNA | [20] |
| Milk microbiome | Streptococcus, Staphylococcus, Serratia and Corynebacteria | V1-V2 16S rRNA | [21] | |
| 7. Adipobiome | Adipose tissue microbiome | Proteobacteria and Firmicutes | V4-V5 16S rRNA | [22] |
| Adipose tissue microbiome related to type 2 diabetes (T2D) and obesity humans | Pseudomonas, Faecalibacterium, Bacteroides and Enterobacter | V3-V4 16S rRNA | [23] | |
| 8. Hepatobiome | Liver microbiome in obese and non-obese humans | Obese: Proteobacteria, Massilia spp. | V3-V4 16S rRNA | [24] |
| Liver tissue microbiome related to diabetes and obesity humans | Obese: Pseudomonas, Arthrobacter and Ruminococcus | V3-V4 16S rRNA | [23] | |
| Liver microbiome in Humans and Mice | Mice: Pseudomonas, Delftia and Coprococcus Humans: Proteobacteria |
16S rRNA | [25] |
2.2. Penilebiome
3. Microbiomes Associated with the Early Life Development
3.1. Amniobiome
3.2. Placentalbiome
3.3. Meconiobiome
3.4. Lactobiome
4. Microbiomes Associated with Later-Life Metabolic Health: Emerging Research Areas
4.1. Adipobiome
4.2. Hepatobiome
References
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103.
- Marchesi, J.R.; Ravel, J. The Vocabulary of Microbiome Research: A Proposal; Springer: Berlin/Heidelberg, Germany, 2015; Volume 3, pp. 1–3.
- Vemuri, R.; Shankar, E.M.; Chieppa, M.; Eri, R.; Kavanagh, K. Beyond just bacteria: Functional biomes in the gut ecosystem including virome, mycobiome, archaeome and helminths. Microorganisms 2020, 8, 483.
- Aagaard, K.; Ma, J.; Antony, K.M.; Ganu, R.; Petrosino, J.; Versalovic, J. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014, 6, 237ra65.
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129.
- Hu, J.; Nomura, Y.; Bashir, A.; Fernandez-Hernandez, H.; Itzkowitz, S.; Pei, Z.; Stone, J.; Loudon, H.; Peter, I. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE 2013, 8, e78257.
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145.
- Roswall, J.; Olsson, L.M.; Kovatcheva-Datchary, P.; Nilsson, S.; Tremaroli, V.; Simon, M.-C.; Kiilerich, P.; Akrami, R.; Krämer, M.; Uhlén, M. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe 2021, 29, 765–776.
- Shukla, G.; Arya, S.; Goyal, P.; Channa, U. Comparative microbial analysis and assessment of paired umbilical cord blood culture and peripheral venous blood culture in neonates with high risk factors to detect early onset neonatal sepsis: A study from tertiary care hospital of central India. Asian J. Med. Sci. 2022, 13, 134–139.
- Vemuri, R.; Sylvia, K.E.; Klein, S.L.; Forster, S.C.; Plebanski, M.; Eri, R.; Flanagan, K.L. The microgenderome revealed: Sex differences in bidirectional interactions between the microbiota, hormones, immunity and disease susceptibility. Semin. Immunopathol. 2019, 41, 265–275.
- Serrano, M.G.; Parikh, H.I.; Brooks, J.P.; Edwards, D.J.; Arodz, T.J.; Edupuganti, L.; Huang, B.; Girerd, P.H.; Bokhari, Y.A.; Bradley, S.P. Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy. Nat. Med. 2019, 25, 1001–1011.
- Fettweis, J.M.; Brooks, J.P.; Serrano, M.G.; Sheth, N.U.; Girerd, P.H.; Edwards, D.J.; Strauss Iii, J.F.; Jefferson, K.K.; Buck, G.A.; Vaginal Microbiome, C. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014, 160, 2272.
- MacIntyre, D.A.; Chandiramani, M.; Lee, Y.S.; Kindinger, L.; Smith, A.; Angelopoulos, N.; Lehne, B.; Arulkumaran, S.; Brown, R.; Teoh, T.G. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci. Rep. 2015, 5, 8988.
- Mehta, S.D.; Zhao, D.; Green, S.J.; Agingu, W.; Otieno, F.; Bhaumik, R.; Bhaumik, D.; Bailey, R.C. The microbiome composition of a man’s penis predicts incident bacterial vaginosis in his female sex partner with high accuracy. Front. Cell. Infect. Microbiol. 2020, 433.
- DiGiulio, D.B.; Callahan, B.J.; McMurdie, P.J.; Costello, E.K.; Lyell, D.J.; Robaczewska, A.; Sun, C.L.; Goltsman, D.S.A.; Wong, R.J.; Shaw, G. Temporal and spatial variation of the human microbiota during pregnancy. Proc. Natl. Acad. Sci. USA 2015, 112, 11060–11065.
- Mehta, S.D.; Nandi, D.; Agingu, W.; Green, S.J.; Bhaumik, D.K.; Bailey, R.C.; Otieno, F. Vaginal and penile microbiome associations with herpes simplex virus type 2 in women and their male sex partners. J. Infect. Dis. 2022, 226, 644–654.
- Kayem, G.; Doloy, A.; Schmitz, T.; Chitrit, Y.; Bouhanna, P.; Carbonne, B.; Jouannic, J.M.; Mandelbrot, L.; Benachi, A.; Azria, E.; et al. Antibiotics for amniotic-fluid colonization by Ureaplasma and/or Mycoplasma spp. to prevent preterm birth: A randomized trial. PLoS ONE 2018, 13, e0206290.
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193.
- Williams, N.; Vella, R.; Zhou, Y.; Gao, H.; Mass, K.; Townsel, C.; Campbell, W.; Luo, G. Investigating the origin of the fetal gut and placenta microbiome in twins. J. Matern. Fetal Neonatal Med. 2021, 35, 7025–7035.
- Pace, R.M.; Williams, J.E.; Robertson, B.; Lackey, K.A.; Meehan, C.L.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W. Variation in human milk composition is related to differences in milk and infant fecal microbial communities. Microorganisms 2021, 9, 1153.
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313.
- Massier, L.; Chakaroun, R.; Tabei, S.; Crane, A.; Didt, K.D.; Fallmann, J.; von Bergen, M.; Haange, S.-B.; Heyne, H.; Stumvoll, M. Adipose tissue derived bacteria are associated with inflammation in obesity and type 2 diabetes. Gut 2020, 69, 1796–1806.
- Anhê, F.F.; Jensen, B.A.H.; Varin, T.V.; Servant, F.; Van Blerk, S.; Richard, D.; Marceau, S.; Surette, M.; Biertho, L.; Lelouvier, B. Type 2 diabetes influences bacterial tissue compartmentalisation in human obesity. Nat. Metab. 2020, 2, 233–242.
- Suppli, M.P.; Bagger, J.I.; Lelouvier, B.; Broha, A.; Demant, M.; Kønig, M.J.; Strandberg, C.; Lund, A.; Vilsbøll, T.; Knop, F.K. Hepatic microbiome in healthy lean and obese humans. JHEP Rep. 2021, 3, 100299.
- Leinwand, J.C.; Paul, B.; Chen, R.; Xu, F.; Sierra, M.A.; Paluru, M.M.; Nanduri, S.; Alcantara, C.G.; Shadaloey, S.A.A.; Yang, F. Intrahepatic microbes govern liver immunity by programming NKT cells. J. Clin. Investig. 2022, 132, e151725.
- Chu, D.M.; Seferovic, M.; Pace, R.M.; Aagaard, K.M. The microbiome in preterm birth. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 52, 103–113.
- Parris, K.M.; Amabebe, E.; Cohen, M.C.; Anumba, D.O. Placental microbial–metabolite profiles and inflammatory mechanisms associated with preterm birth. J. Clin. Pathol. 2021, 74, 10–18.
- Olaniyi, K.S.; Moodley, J.; Mahabeer, Y.; Mackraj, I. Placental microbial colonization and its association with pre-eclampsia. Front. Cell. Infect. Microbiol. 2020, 10, 413.
- Singh, A.; Mittal, M. Neonatal microbiome–a brief review. J. Matern. Fetal Neonatal Med. 2020, 33, 3841–3848.
- Lundgren, P.; Thaiss, C.A. The microbiome-adipose tissue axis in systemic metabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G717–G724.
- Tilg, H.; Burcelin, R.; Tremaroli, V. Liver tissue microbiome in NAFLD: Next step in understanding the gut–liver axis? Gut 2020, 69, 1373–1374.
