Aptamers are single-stranded DNA or RNA sequences of 20-80 nucleotides that interact with different targets such as: proteins, ions, viruses, or toxins, through non-covalent interactions and their unique three-dimensional conformation. They are obtained in vitro by the systematic evolution of ligands by exponential enrichment (SELEX). Because of their ability of target recognition with high specificity and affinity, aptamers are usually compared to antibodies. However, they present many advantages that make them promising molecules for the development of new methods for the diagnosis and treatment of human diseases. In medical parasitology, aptamers also represent an attractive alternative for the implementation of new parasite detection methods, easy to apply in endemic regions. The aim of this study was to describe the current advances in the development of diagnostic tests based on aptamers in parasitology. For this, articles were selected following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, with specific inclusion and exclusion criteria. The 26 resulting articles deal with the use of aptamers for the detection of six important protozoa that affect human health. This systematic review clearly demonstrates the specificity, sensitivity and selectivity of aptamers and aptasensors, that certainly will soon become standard methods in medical parasitology.
- aptamers
- detection test
- diagnosis
- parasites
- SELEX
1. Introduction
Aptamers were developed for the first time in 1990 by two different research groups [1][2][1,2]. They are single stranded (ss) DNA or RNA molecules of approximately 20–80 nucleotides that recognize and interact with different targets including proteins [3], ions [4], viruses [5], and toxins [6], among others, through no-covalent interactions like van der Waals forces, hydrogen bonds, electrostatic interactions and by complementary forms, due to their unique three-dimensional conformation. Aptamers are usually designed from an in vitro iterative technique named systematic evolution of ligands by exponential enrichment (SELEX) [1][2][1,2], through several rounds of selection (seven to ten) that allow one to obtain molecules with high specificity and affinity for the target. Notably, the dissociation constants of aptamers are usually in the nano/picomolar range, which is comparable to those reported for antibodies [7].
Due to their ability of recognizing specific targets, aptamers are often compared to antibodies. However, they present several advantages over them. First, aptamers are generated by chemical synthesis without batch-to-batch variation, ensuring the constant functionality of the molecules. Another advantage is related to the production time, which is relatively short for aptamers (less than three months) compared with the longer production period required for monoclonal antibodies (approximately six months); consequently, the synthesis of aptamers is less expensive, whereas the use of cells and animal models largely increases the cost of antibodies [8]. Moreover, even though aptamers can be denatured, they can return to their original conformation and functionality after the optimal temperature is restored; this characteristic is particularly important since it allows prolonged storage periods and transportation at room temperature, which is not possible with antibodies. Furthermore, aptamers are 10–100 times smaller than antibodies, allowing them to penetrate more easily into the tissues [9]; they can also be modified at their 5′ or 3′ ends, or in their sugar-phosphate chain, to prevent degradation by nucleases or reduce renal filtration, which enhances their stability in vivo and their capability to be used in therapy [10]. Finally, aptamers can be conjugated with biotin, digoxigenin, fluorescent markers, and others—without losing their target affinity and specificity—to be used in diagnostic techniques such as flow cytometry [11], biosensors [12][13][14][12–14], enzyme-linked aptamer-based apta-sorbent assay (ELASA) [15], and other multiple applications.
To date, aptamers represent a potential alternative for the diagnosis of cardiovascular diseases [16], cancer [17], and infectious diseases [18][19][18,19]. One example is the aptasensor developed by Negahdary and coworkers [16] to detect the cardiac troponin I (TnI) for the early diagnosis of acute myocardial infarction. Results showed a specificity of 81% and a sensitivity of 100% with a detection limit of 10 pg/mL which is better than the conventional enzyme linked immunosorbent assay (ELISA) kit currently employed, indicating that this aptasensor could be useful for the diagnosis of myocardial infarction. Another example is the work of Xi and colleagues [19] that describes the design of an aptamer that recognizes the surface antigen of hepatitis B virus (HBsAg) and can be used in a quimioluminescent aptasensor for hepatitis B diagnostic, in order to replace the conventional ELISA that is a more complex technique and requires a longer processing time; importantly, the aptasensor has a detection limit of 0.05 ng/mL, which is 10-fold lower than the value reported for the ELISA method (0.5 ng/mL).
2. Article Selection
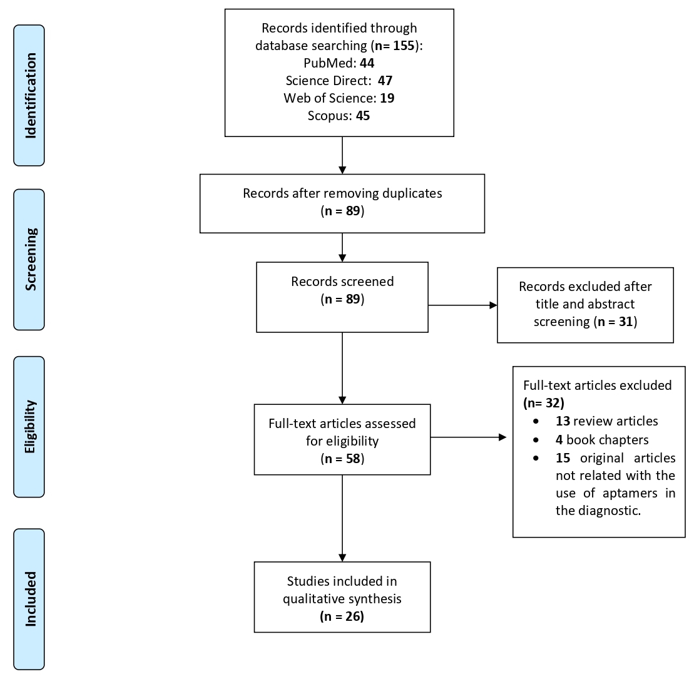
A total of 155 articles were obtained from the primary searches, of which 44 were from PubMed, 47 from ScienceDirect, 19 from Web of Science and 45 from Scopus. A group of 66 duplicated papers were removed, leaving a total of 89 articles. Then, we performed a filter step based on the title, abstract and keywords, which reduced the selection to 58 studies. Next, the full text of these 58 articles was reviewed and 32 were excluded because: 13 were review articles, four were book chapters and 15 were not studies related with diagnosis methods for parasite infections. Finally, only 26 articles that met all the inclusion criteria were included in this systematic review (Figure 1). The selected articles are focused on the use of aptamers for the detection of protozoan parasites that affect human health, particularly Plasmodium, Leishmania, Trypanosoma, Cryptosporidium, Toxoplasma, and Trichomonas.
Figure 1. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram for article selection included in this systematic review.
3. Plasmodium spp.
Plasmodium spp. is responsible for malaria, the first leading cause of death from parasites, with 228 million cases and approximately 400,000 deaths each year [21]. Malaria can be cured if detected early, which highlights the need for efficient and easy to use diagnostic methods. Currently, microscopy detection, with or without fluorescent dyes to increase the sensitivity, remains one of the principal methods for malaria diagnosis, although the need for laboratory equipment can be difficult in the field. The use of the polymerase chain reaction (PCR), and colorimetric assays for Plasmodium detection is limited by the need of experienced laboratory personnel and considerable sophisticated equipment, while kits based on immobilized labelling antibodies are not authorized for clinical use yet [22]. These observations clearly indicate that better rapid diagnostic tests (RDTs) of malaria are required. Notably, three main groups of investigation have focused their efforts on the development of simple and rapid aptamer-based diagnosis methods for malaria (Table 1).
3.1. Aptasensors Using the pL1 Aptamer to Detect the Parasite Lactate Dehydrogenase
A first set of aptasensors for Plasmodium detection were designed using ssDNA aptamers raised against the parasite lactate dehydrogenase (LDH) known as an important diagnostic target for malaria. Three of the selected articles used the pL1 DNA aptamer (also called pLDH), obtained by the group of Ban after 10 rounds of selection in a standard SELEX method, based on His-tag P. vivax LDH (PvLDH) proteins immobilized on magnetic beads. Interestingly, the pL1 aptamer recognized both PvLDH and P. falciparum LDH (PfLDH) with Kd values of 16.8 ± 0.6 nM and 38.7 ± 1.3 nM, respectively [23]. In the first strategy, the same group took advantage of the property of two water-soluble cationic polymers, poly(diallyldimethylammonium chloride) (PDDA) and poly(allylamine hydrochloride) (PAH) to assemble DNA molecules, i.e., pL1 aptamers, into nanostructures, and aggregate gold-nanoparticles (AuNPs) that have unique optical and electronic properties. The method was based on the competitive binding of PDDA or PAH to the pL1 aptamer and AuNPs, depending on the presence or absence of LDH. In the absence of LDH, cationic polymers are sequestered by free pL1 aptamers and thus, cannot aggregate AuNPs. In contrast, the formation of the pL1-LDH complex allows them to accumulate AuNPs, promoting a color change from red to blue that can be easily measured and quantified by using a UV-Vis spectrophotometer and transmission electron microscopy (TEM). After optimization (3.5 nM PDDA or 25 nM PAH, and 10 nM of the pL1 aptamer), the aptasensors were able to detect from 8.7 pM PvLDH for PDDA and 8.3 pM PvLDH for PAH, and 10.3 pM PfLDH for PDDA and 12.5 pM PfLDH for PAH. They were highly selective towards recombinant parasite proteins since they did not detect lysozyme and bovine serum albumin (BSA) that were used as competitive proteins. Importantly, they also permitted the detection of 80 P. vivax parasites/μL (92 P. falciparum parasites/μL) for PDDA and 74 P. vivax parasites/μL (97 P. falciparum parasites/μL) for PAH [24]. Then, the same authors showed that the cationic surfactant hexadecyltrimethylammonium bromide (CTAB) is a better candidate for their pL1-based aptasensor, since 4 nM of CTAB with 20 nM of the pL1 aptamer was able to detect from 1.25 pM PvLDH and 2.94 pM PfLDH. This biosensor also identified Plasmodium LDH in human serum with limits of detection (LOD) of 10.17 pM and 13.54 pM for PvLDH and PfLDH, respectively, indicating that this simple, sensitive, and selective colorimetric biosensor has a great potential for the diagnosis of malaria [25].
The pL1-aptamer was also used by Geldert and colleagues to develop an interesting fluorescence resonance energy transfer (FRET)-based paper aptasensor. Aptamers were adsorbed on MoS2 nanosheets through the fluorescein amidites (FAM) labeling at their 3′-end. After studying the wettability, and the micro- and nanoscale surface morphologies of five types of paper (two chromatography papers, Whatman Qualitative Filter Paper (No. 1) and Advantec Chromatography Paper (No. 51B), Lens paper (grade 541, VWR, Radnor, PA, USA), A4 printer paper (double A) and coffee filters (Boncafe)), the authors verified that labeled aptamers-MoS2 were distributed throughout the microstructure of the paper and that fluorescence quenching could be quantified from samples dried on paper, as it can be done in classical liquid-based FRET sensors. Finally, strips of paper previously coated with aptamers-MoS2 nanosheets (100 nM pL1 aptamers and 25 μg/mL MoS2 nanosheets) were tested for their reactivity with recombinant P. falciparum LDH and unrelated proteins. Surprisingly, printer paper was the only material that showed a significant and specific production of fluorescence on the strip after 30 min of incubation, when aptamers were released from the nanosheet quenchers to bind the parasite LDH (30 nM), making it the most appropriate substrate for this paper-based FRET diagnosis system. This could be due to its lower wettability and smaller pore size, which might maintain the interactions with aptamers-MoS2, allowing for better binding with target molecules. This rational strategy established the basis of a facile, disposable, and low-cost diagnostic FRET-based paper aptasensor that could be easily applied in low-resource areas where infectious diseases such as malaria are endemic [26].
3.2. Aptasensors Using the 2008a Aptamer to Detect the Parasite Lactate Dehydrogenase
The group of Tanner developed several bisosensors based on another ssDNA aptamer recognizing the PfLDH, named as 2008s, that was selected after 20 rounds of SELEX using PfLDH immobilized on magnetic beads, and the human homologue for the elimination of nonspecific aptamers. After coupling with AuNPs via a 5′ thiol group, the interaction between PfLDH and 2008s aptamer was determined by isothermal titration calorimetry (ITC), surface plasmon resonance (SPR) and electrophoretic mobility shift assay (EMSA), and showed Kd values of 42 nM, 59 nM and 56 ± 18 nM, respectively. Methods such as TEM evidenced the aggregation of 2008s-AuNPs in the presence of PfLDH. In addition, the loss of the red color was evidenced in the presence of PfLDH in comparison with the observed color in the presence of the human homologue. Interestingly, the LOD of this method was 57 pg/μL PfLDH, therefore this colorimetric assay could be used for the detection of PfLDH in the plasma (2–15 pg/ μL) of patients with malaria [27].
The fact that binding of 2008s to PfLDH does not affect the enzyme activity, prompted the group of Tanner to use 2008s in an aptamer-tethered enzyme capture (APTEC) system able to detect PfLDH in blood samples. For this, lysed blood samples were incubated in 2008s-coated wells to allow PfLDH binding; after washing, LDH activity was easily quantified by the standard colorimetric assay using L-lactate as a substrate and the nitrotetrazolium blue chloride (NTB) dye. The LOD were calculated as 4.9 ng/mL ± 2 ng/mL PfLDH in blood samples, as well as 600 ± 250 parasites/mL for asynchronous parasite culture, and 3500 ± 250 parasites/ mL for ring stage parasites. A comparative assay evidenced that the APTEC test is as efficient as the antibody-based OptiMAL-IT dipsticks to diagnose P. falciparum infections in patient blood samples. Moreover, 2008s-coated wells remained able to bind PfLDH in at least six successive APTEC assays, which may allow large population analysis at a lower cost. The authors also demonstrated that it is possible to perform a semi-quantitative APTEC assay by varying the aptamer density to determine PfLDH concentration by visual assessment, which makes it a promising tool to follow parasitemia levels in patients [28]. Additionally, using the same APTEC strategy, Cheung and coworkers in 2018 showed that aptamer 2008s only exhibits a slight signal for P. vivax LDH, due to the recognition of a similar cofactor-binding cleft in the enzyme of both Plasmodium species, but it did not recognize human LDH and serum albumin, confirming the specificity of 2008s for PfLDH. A remarkable finding of this work is the recognition of the target protein directly in blood samples of infected patients. However, the microtiter plate format limits the general application of this APTEC system in endemic zones, in which the use of electrochemical sensors is generally preferred [29]. Therefore, the same group developed a 3D printed microfluidic biosensor that could be easily used in the field. Briefly, magnetic beads (80 μg) coated with biotinylated 2008s aptamers were incubated with lysed sample of human blood in the incubation chambers, then they were magnet-guided to the wash chamber, and finally, magnetic bead-aptamer-PfLDH complexes were revealed by the formation of an insoluble purple colored formazan dye in the development chamber, that can be captured by a standard mobile-cellular camera phone. This portable biosensor was about 14-fold more sensitive that the original APTEC assay since it allowed for the detection of at least 250 ring stage parasites/μL; moreover, it was specific for P. falciparum, which makes it a very promising candidate for a rapid diagnostic test [30].
The 2008s aptamer was also used to develop an original aptasensor based on DNA origami technology. The 5’-end of the 2008s aptamer was ligated to a polyT linker sequence followed by a staple strand that is complementary to 12 different regions of the M13 plasmid sequence. Interestingly, these staple-polyT-aptamers seemed to have a greater affinity for PfLDH that the original 2008s sequence, with higher apparent Kd values from 1090 ± 183 to 647 ± 128 nM in EMSA. Then, modified aptamers were mixed with M13 DNA to form a rectangular DNA origami with at least one aptamer protruding from the structure. Atomic force microscopy imaging revealed that a maximum number of four pfLDH are bound to the DNA origami, and this is probably regulated by the size of the protein and its interaction with two aptamers. Consistent with EMSA data, the LOD was found at 500 nM PfLDH. The DNA origami aptasensor was still able to recognize PfLDH mixed in blood plasma, which is a necessary requirement for diagnosis; moreover, the fact that bound LDH still has enzymatic activity may promote the use of fluorescent molecules to easily visualize the presence of PfLDH in infected samples [31].
Figueroa-Miranda, of the Mayer group, decided to immobilize the 2008s aptamer (0.5 µM) on a gold electrode to develop an electrochemical impedance aptasensor. First, they used 6-mercapto-1-hexanol (6-MCH) as a spacer molecule to block empty spaces, avoid unspecific interactions and allow an accurate interaction between the aptamer and PfLDH. The LOD of the 6-MCH-aptasensor was 0.84 pM PfLDH, which is comparable with other methods previously described for the diagnostic of malaria. The aptasensor also detected the PfLDH in 10-fold diluted human serum with a low LOD of 1.3 pM and a dynamic detection range from 10 pM to 10 nM, indicating that it could be useful for the early stage of P. falciparum infection. Moreover, its specificity was established by the use of unrelated proteins, such as bovine and human serum albumin, as well as mouse and human LDH. Interestingly, this 6-MCH-aptasensor can be regenerated with 6 M urea and kept at 4 °C without affecting the binding of the aptamer to the electrode and its affinity for PfLDH, which may reduce costs and facilitates its use in the field [32]. Later, the same group reported improvement of the original design by replacing 6-MCH by polyethylene glycol (PEG). Interestingly, the LOD was reduced to 0.8 pM PfLDH in two-fold diluted human serum, and the detection range was increased (2.3 pM–100 nM). On the other hand, the results of ex situ experiments showed that the PEG-aptasensor incubated with whole human serum presented a LOD of 1.49 pM, with a detection range of 4.5 pM–100 nM, that agrees with the results of in situ experiments. Similarly, as other aptasensors, the PEG-aptasensor only recognized PfLDH but not other proteins present in serum. Considering all the aforementioned information, this aptasensor is an interesting in situ tool that could be used in point-of-care for PfLDH detection [33].
Finally, Kim and Searson used the 2008s aptamer in an aptabiosensor based on aptamer modified magnetic microparticles (MMP) for capture, oligonucleotide-modified quantum dots (QD) for detection, and oligonucleotide-modified AuNPs for signal amplification. First, through a half-sandwich assay using QD conjugated to a cOligos aptamer-cOligo-QD construct (1:6 molar ratio), they proved the recognition of the PfLDH and PvLDH proteins immobilized on a 96-well plate. The next step included the integration of MPP functionalized with aptamers (1:6 ratio) in a MMP-QD sandwich assay which allowed the detection of 0.033–3 ng of PfLDH and PvLDH (0.51 to 46 fmole). Then, for signal amplification and to reduce the LOD, a MMP-AuNP-QD aptamer sandwich assay was designed. Results showed a considerable reduction in the LOD down to 0.66 pg PfLDH and PvLDH proteins, demonstrating a 50-fold higher sensitivity than the MMP-QD aptamer sandwich assay. However, the complete validation of this method still requires the evaluation of clinical samples from patients infected with Plasmodium [34].
3.3. Aptasensors Using the P38 Aptamer to Detect the Parasite Lactate Dehydrogenase
In 2016, the PfLDH protein was used by the group of Goswami to obtain the P38 ssDNA aptamer after 10 rounds of SELEX selection with the parasite protein immobilized on a polyvinylidene difluoride (PVDF) membrane, as well as human LDH chain A, human LDH chain B and protein-free PVDF membrane for negative selection steps. EMSA study confirmed the binding affinity of P38, showing a Kd value of 0.35 µM. The P38 aptamer was employed for the standardization of an aptasensor based on the use of graphene oxide (GO) as an immobilization matrix. The specificity of the aptasensor for PfLDH protein was confirmed voltammetrically (0.65 V) by measuring the NADH generated from the oxidation of lactate by PfLDH. The P38-aptasensor has other interesting properties, such as an LOD of 0.5 fM with a detection range of 0.5 fM–10 fM; it is able to detect PfLDH in mimicked real samples spiked with 5 fM of recombinant PfLDH, while the sensitivity of most aptasensors generally is in the picomolar range. Furthermore, this aptasensor was able to evidence parasitemia in 20-fold diluted whole samples of patients with P. vivax infection, whereas the commercial malaria kits based on antibody based LDH were not. For these reasons the authors propose the use of this aptasensor for samples with a low parasitemia [35].
3.4. Aptasensors Using the NG3 Aptamer to Detect the Parasite Glutamate Dehydrogenase
The other parasite biomarker selected for aptamer design is the P. falciparum glutamate dehydrogenase (PfGDH) that can be found in patient blood during the sexual and asexual stages of the parasite development. Recently, the group of Goswami obtained the ssDNA NG3 aptamer from 17 rounds of SELEX using the PfGDH immobilized on a PVDF membrane, with additional cycles using a protein-free PVDF membrane and human GDH, respectively, to discard non-specific molecules. In an SPR study, the thiolated NG3 was able to specifically bind to the PfGDH antigen with high affinity (Kd = 79 nM). A monolayer of NG3 aptamer was then chemically immobilized over gold disc electrodes and non-Faradaic electrochemical impedance spectroscopy measurements showed that the aptasensor produced a capacitance response at an optimized frequency of 2 Hz following its binding with the target PfGDH. The LOD was found to be 0.43 pM and 0.77 pM for PfGDH in buffer and in serum, respectively, without significant detection of other malarial biomarkers (PfLDH and PfHRP-II), which makes it a high performance aptasensor for malaria diagnosis [36]. The same group also immobilized a self-assembled monolayer of NG3 aptamer on the surface of an inter-digitated gold microelectrode (IDµE) connected to the gate of a field effect transistor (FET), to develop a miniaturized aptamer-based FET biosensor that exhibited a specific and sensitive response to PfGDH in spiked buffer and serum samples, with an LOD of 16.7 pM and 48.6 pM, respectively, which may allow the easy diagnosis of symptomatic and asymptomatic patients with low parasitemia levels in the field [37].
Finally, in a recent study, the same authors used biotinylated P38 and NG3 aptamers to coat magnetic beads (200 μg) to capture Plasmodium LDH and PfGDH, respectively, and quantitatively detected them using a cocktail buffer containing lactate and glutamate as substrates, through the conversion of the resazurin dye to resorufin at 600 nm. Data of absorbance and fluorescence intensity revealed that LOD values were 0.55 ± 0.09 pM and 1.72 ± 0.13 pM, respectively, for pan LDH in human blood serum, whereas they were 1.34 ± 0.12 and 1.43 ± 0.14 pM, respectively, in the case of serum spiked with PfGDH. Alternatively, the enzyme reaction was transferred to a Diethylaminoethyl (DEAE)-cellulose surface modified (DSM) chromatographic paper in order to develop an instrument-free portable test based on the use of a standard camera and the ImageJ software for densitometry analysis of the developed color. Results showed that the LOD for pan LDH and PfGDH in the serum sample were 69.25 ± 8.22 pM and 68.75 ± 7.64 pM, respectively. This high sensitivity and the absence of reaction with nonspecific proteins, indicate the potential of this aptasensor for malaria diagnosis [38].
References
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510.
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822.
- Ohuchi, S.; Mori, Y.; Nakamura, Y. Evolution of an Inhibitory RNA Aptamer against T7 RNA Polymerase. FEBS Open Bio 2012, 2, 203–207.
- Raducanu, V.-S.; Rashid, F.; Zaher, M.S.; Li, Y.; Merzaban, J.S.; Hamdan, S.M. A Direct Fluorescent Signal Transducer Embedded in a DNA Aptamer Paves the Way for Versatile Metal-Ion Detection. Actuators B Chem. 2020, 304, 127376. https://doi.org/10.1016/j.snb.2019.127376
- Shiratori, I.; Akitomi, J.; Boltz, D.A.; Horii, K.; Furuichi, M.; Waga, I. Selection of DNA Aptamers That Bind to Influenza A Viruses with High Affinity and Broad Subtype Specificity. Biophys. Res. Commun. 2014, 443, 37–41.
- Liu, F.; Ding, A.; Zheng, J.; Chen, J.; Wang, B. A Label-Free Aptasensor for Ochratoxin a Detection Based on the Structure Switch of Aptamer. Sensors 2018, 18, 1769.
- Aptekar, S.; Arora, M.; Lawrence, C.L.; Lea, R.W.; Ashton, K.; Dawson, T.; Alder, J.E.; Shaw, L. Selective Targeting to Glioma with Nucleic Acid Aptamers. PLoS ONE 2015, 10, e0134957. https://doi.org/10.1371/journal.pone.0134957
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. J. Mol. Sci. 2017, 18, 2142. doi: 10.3390/ijms18102142
- Xiang, D.; Zheng, C.; Zhou, S.-F.; Qiao, S.; Tran, P.H.-L.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics 2015, 5, 1083–1097.
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a Targeted Anti-VEGF Aptamer for Ocular Vascular Disease. Rev. Drug Discov. 2006, 5, 123–132.
- Zhang, P.; Zhao, N.; Zeng, Z.; Feng, Y.; Tung, C.-H.; Chang, C.-C.; Zu, Y. Using an RNA Aptamer Probe for Flow Cytometry Detection of CD30-Expressing Lymphoma Cells. Investig. 2009, 89, 1423–1432.
- Eissa, S.; Zourob, M. Aptamer-Based Label-Free Electrochemical Biosensor Array for the Detection of Total and Glycated Hemoglobin in Human Whole Blood. Rep. 2017, 7, 1016. https://doi.org/10.1038/s41598-017-01226-0
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Chem. Int. Ed. 2017, 56, 11916–11920.
- Srivastava, M.; Nirala, N.R.; Srivastava, S.K.; Prakash, R. A Comparative Study of Aptasensor Vs Immunosensor for Label-Free PSA Cancer Detection on GQDs-AuNRs Modified Screen-Printed Electrodes. Rep. 2018, 8, 1923. https://doi.org/10.1038/s41598-018-19733-z
- Li, P.; Zhou, L.; Wei, J.; Yu, Y.; Yang, M.; Wei, S.; Qin, Q. Development and Characterization of Aptamer-Based Enzyme-Linked Apta-Sorbent Assay for the Detection of Singapore Grouper Iridovirus Infection. Appl. Microbiol. 2016, 121, 634–643.
- Negahdary, M.; Behjati-Ardakani, M.; Sattarahmady, N.; Heli, H. An Aptamer-Based Biosensor for Troponin I Detection in Diagnosis of Myocardial Infarction. Biomed. Phys. Eng. 2018, 8, 167–178.
- Pan, L.; Zhao, J.; Huang, Y.; Zhao, S.; Liu, Y.-M. Aptamer-Based Microchip Electrophoresis Assays for Amplification Detection of Carcinoembryonic Antigen. Chim. Acta 2015, 450, 304–309.
- Kim, D.T.H.; Bao, D.T.; Park, H.; Ngoc, N.M.; Yeo, S.-J. Development of a Novel Peptide Aptamer-Based Immunoassay to Detect Zika Virus in Serum and Urine. Theranostics 2018, 8, 3629–3642.
- Xi, Z.; Gong, Q.; Wang, C.; Zheng, B. Highly Sensitive Chemiluminescent Aptasensor for Detecting HBV Infection Based on Rapid Magnetic Separation and Double-Functionalized Gold Nanoparticles. Rep. 2018, 8, 9444. https://doi.org/10.1038/s41598-018-27792-5
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. https://doi.org/10.1371/journal.pmed.1000097
- World Health Organization. Available online: https://www.who.int/malaria/publications/world-malaria-report-2015/report/en/ (accessed on 15 August 2020).
- Cunningham, J.; Jones, S.; Gatton, M.L.; Barnwell, J.W.; Cheng, Q.; Chiodini, P.L.; Glenn, J.; Incardona, S.; Kosack, C.; Luchavez, J.; et al. A review of the WHO malaria rapid diagnostic test product testing programme (2008–2018): Performance, procurement and policy. J. 2019, 18, 387. https://doi.org/10.1186/s12936-019-3028-z
- Lee, S.; Song, K.-M.; Jeon, W.; Jo, H.; Shim, Y.-B.; Ban, C. A Highly Sensitive Aptasensor towards Plasmodium Lactate Dehydrogenase for the Diagnosis of Malaria. Bioelectron. 2012, 35, 291–296.
- Jeon, W.; Lee, S.; Dh, M.; Ban, C. A Colorimetric Aptasensor for the Diagnosis of Malaria Based on Cationic Polymers and Gold Nanoparticles. Biochem. 2013, 439, 11–16.
- Lee, S.; Manjunatha, D.H.; Jeon, W.; Ban, C. Cationic Surfactant-Based Colorimetric Detection of Plasmodium Lactate Dehydrogenase, a Biomarker for Malaria, Using the Specific DNA Aptamer. PLoS ONE 2014, 9, e1 https://doi.org/10.1371/journal.pone.0100847
- Geldert, A.; Lim, C.T. Paper-Based MoS2 Nanosheet-Mediated FRET Aptasensor for Rapid Malaria Diagnosis. Rep. 2017, 7, 17510. doi: 10.1038/s41598-017-17616-3.
- Cheung, Y.-W.; Kwok, J.; Law, A.W.L.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural Basis for Discriminatory Recognition of Plasmodium Lactate Dehydrogenase by a DNA Aptamer. Natl. Acad. Sci. USA 2013, 110, 15967–15972.
- Dirkzwager, R.M.; Kinghorn, A.B.; Richards, J.S.; Tanner, J.A. APTEC: Aptamer-Tethered Enzyme Capture as a Novel Rapid Diagnostic Test for Malaria. Commun. 2015, 51, 4697–4700.
- Cheung, Y.-W.; Dirkzwager, R.M.; Wong, W.-C.; Cardoso, J.; D’Arc Neves Costa, J.; Tanner, J.A. Aptamer-Mediated Plasmodium-Specific Diagnosis of Malaria. Biochimie 2018, 145, 131–136.
- Fraser, L.A.; Kinghorn, A.B.; Dirkzwager, R.M.; Liang, S.; Cheung, Y.-W.; Lim, B.; Shiu, S.C.-C.; Tang, M.S.L.; Andrew, D.; Manitta, J.; et al. A Portable Microfluidic Aptamer-Tethered Enzyme Capture (APTEC) Biosensor for Malaria Diagnosis. Bioelectron. 2018, 100, 591–596.
- Godonoga, M.; Lin, T.-Y.; Oshima, A.; Sumitomo, K.; Tang, M.S.L.; Cheung, Y.-W.; Kinghorn, A.B.; Dirkzwager, R.M.; Zhou, C.; Kuzuya, A.; et al. A DNA Aptamer Recognising a Malaria Protein Biomarker Can Function as Part of a DNA Origami Assembly. Rep. 2016, 6, 21266. doi: 10.1038/srep21266
- Figueroa-Miranda, G.; Feng, L.; Shiu, S.C.-C.; Dirkzwager, R.M.; Cheung, Y.-W.; Tanner, J.A.; Schöning, M.J.; Offenhäusser, A.; Mayer, D. Aptamer-Based Electrochemical Biosensor for Highly Sensitive and Selective Malaria Detection with Adjustable Dynamic Response Range and Reusability. Actuators B Chem. 2018, 255, 235–243.
- Figueroa-Miranda, G.; Wu, C.; Zhang, Y.; Nörbel, L.; Lo, Y.; Tanner, J.A.; Elling, L.; Offenhäusser, A.; Mayer, D. Polyethylene Glycol-Mediated Blocking and Monolayer Morphology of an Electrochemical Aptasensor for Malaria Biomarker Detection in Human Serum. Bioelectrochemistry 2020, 136, 107589. https://doi.org/10.1016/j.bioelechem.2020.107589
- Kim, C.; Searson, P.C. Detection of Plasmodium Lactate Dehydrogenase Antigen in Buffer Using Aptamer-Modified Magnetic Microparticles for Capture, Oligonucleotide-Modified Quantum Dots for Detection, and Oligonucleotide-Modified Gold Nanoparticles for Signal Amplification. Chem. 2017, 28, 2230–2234.
- Jain, P.; Das, S.; Chakma, B.; Goswami, P. Aptamer-Graphene Oxide for Highly Sensitive Dual Electrochemical Detection of Plasmodium Lactate Dehydrogenase. Biochem. 2016, 514, 32–37.
- Singh, N.K.; Arya, S.K.; Estrela, P.; Goswami, P. Capacitive Malaria Aptasensor Using Plasmodium Falciparum Glutamate Dehydrogenase as Target Antigen in Undiluted Human Serum. Bioelectron. 2018, 117, 246–252.
- Singh, N.K.; Thungon, P.D.; Estrela, P.; Goswami, P. Development of an Aptamer-Based Field Effect Transistor Biosensor for Quantitative Detection of Plasmodium Falciparum Glutamate Dehydrogenase in Serum Samples. Bioelectron. 2019, 123, 30–35.
- Singh, N.K.; Jain, P.; Das, S.; Goswami, P. Dye Coupled Aptamer-Captured Enzyme Catalyzed Reaction for Detection of Pan Malaria and P. Falciparum Species in Laboratory Settings and Instrument-Free Paper-Based Platform. Chem. 2019, 91, 4213–4422.