Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Christophe Hano.
Flavonoids are secondary metabolites that represent a heterogeneous family of plant polyphenolic compounds. The research has determined that the health benefits of fruits and vegetables, as well as the therapeutic potential of medicinal plants, are based on the presence of various bioactive natural products, including a high proportion of flavonoids. In plant metabolite research, flavonoids have become the center of attention due to their significant bioactivity associated with anti-cancer, antioxidant, anti-inflammatory, and anti-microbial activities.
- flavonoids
- biosynthesis
- metabolic engineering
- microbial production
1. Introduction
Flavonoids comprise an essential and diverse class of polyphenolic compounds that are synthesized in plants as bioactive secondary metabolites, consisting of more than 6000 classified phenolics [1,2,3,4][1][2][3][4]. The synthesis of these secondary metabolites in plants is tissue-specific and highly regulated [5]. They contain 15 carbon atoms and two phenyl rings linked together with a 3-carbonated heterocyclic ring, forming a C6-C3-C6 structure [6,7,8,9][6][7][8][9]. Due to variations in the structure of the backbone, an enormous level of chemical diversity is observed in flavonoids [10]. Based on these variations, the flavonoids are organized into six main groups, namely: flavan-3-ols, flavanones, flavones, flavanols, anthocyanidins, and isoflavones [1,4,6,10][1][4][6][10]. These natural compounds are generally found in citrus grain seeds, vegetables, fruits, nuts, chocolate, cocoa, legumes, black tea, red wine, green tea, soy, herbs, onions, berries, apple, grapes, and cider [3,6,7,8,11][3][6][7][8][11]. In plants, flavonoids are responsible for protecting against oxidative stress, by acting as free radical scavengers, and provide coloration to flowers, leaves, and fruits, hence attracting pollinators. Moreover, these phytochemicals act as UV filters and protect plants from damage caused by UV radiation, function as signaling molecules, serve as antimicrobial defensive compounds, and act as chelators for toxic heavy metals [6,10,12,13][6][10][12][13]. Flavonoid compounds have been reported to have a wide range of potential biological applications in humans, including antibacterial [14], antifungal [8], antiviral [15], anticancer [1], cardioprotective [6], anti-inflammatory [16], antidiabetic [3], antiaging [17], and radioprotective activities [9].
The current methods of producing flavonoids are largely dependent on plant extraction, which has several limitations and drawbacks. Firstly, many plants rich in flavonoids also act as a food source. Hence, their consumption for the production of flavonoids may result in depletion of the overall food supply [18,19][18][19]. Secondly, extraction yields are limited due to low concentrations or loses during extraction; moreover, long breeding seasons, unfavorable climate conditions, small plantation sizes, cultivation difficulties, and variation in plant species also effect production [5,18,20][5][18][20]. Simultaneously, the flavonoid concentration in these plants is comparatively low, limiting their large-scale production [18]. An alternative approach could be chemical synthesis, but due to the complexity of some flavonoid structures, this synthesis approach is expensive and time-consuming [18]. Moreover, the use of potentially hazardous chemicals and intense reaction conditions also restrict the de novo synthesis of these plant secondary metabolic compounds [5,20,21][5][20][21]. As a result, these factors add up to a costly, energy-intensive preparation process with room for further improvement as well [18]. In this context, the reconstitution of biosynthetic gene fragments in plants and industrially important microorganisms holds a lot of promise for flavonoid exploration and scalable production [11,22][11][22]. Plant metabolic engineering promises to increase the production of specific beneficial flavonoids and to expand the chemical range of novel flavonoids [11[11][23],23], but still, it is difficult and technically challenging to engineer a plant genome. The editing of a plant genome for enhanced metabolite production often results in several challenges and limitations in terms of titer variability, aggregation susceptibility, growth rate, stress, and heterogeneity of culture [24]. The various methods of synthesizing flavonoids are depicted in Figure 1.
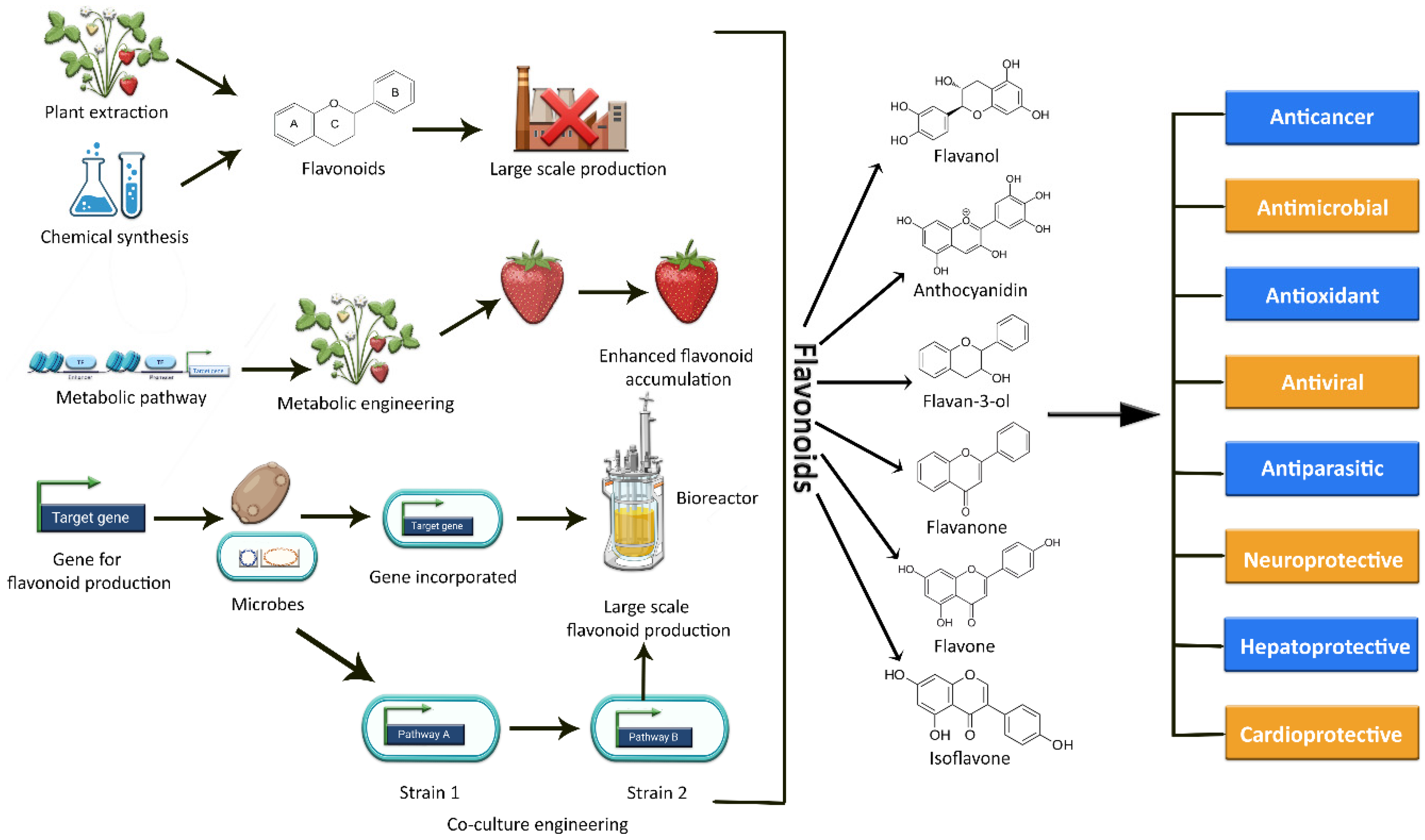
Figure 1. Graphical representation of the various methods used to synthesize a variety of flavonoids, as well as their potential biological activities.
Recently, the microbial production of flavonoids has received ample attention due to its low energy requirements, high purity of the product, and low emissions of waste, such as sulfates, nitrates, or nitrites [20]. Synthetic and system biology technologies have revolutionized the field of metabolic engineering over the past two decades, allowing for the establishment of bio-based production in numerous engineered microbes [25,26][25][26]. This approach has several benefits, such as a rapid growth rate, safety, economic substrates, ease of culture, range of various genetic techniques, and high concentration of desired metabolites as compared to the native host [24]. In recent years, apart from Escherichia coli, a wide array of host strains, including Lactococcus lactis, Yarrowia lipolytica, and Saccharomyces cerevisiae, have been tested for flavonoid production [11]. Nowadays, a different approach involving modular co-culture engineering has surfaced to overcome the constraints of the conventional monoculture approach by harnessing the power of two or more strains of engineered microbes to reconstruct the target biosynthetic pathway. In this way, it has largely reduced the biosynthesis labor, as well as the associated metabolic burden on each microbial strain [27].
2. Flavonoid Biosynthetic Pathways in Plants
There are two distant biosynthetic pathways in plants for the production of flavonoid compounds, e.g., the acetate pathway and the shikimic acid pathway. Initially, the acetate pathway that forms ring A (of flavonoids) and the shikimate pathway that forms ring B (of flavonoids) merge together through a linking chain called ring C (generating the C6-C3-C6 structural components or building-blocks of the flavonoid skeleton) leading to the biosynthesis of flavonoids. The transformation of glucose leads to the synthesis of three molecules of malonyl-CoA from which ring A is synthesized, whereas the shikimate pathway generates the phenyl propanoids through the production of phenylalanine, which gives 4-coumaroyl-CoA molecules as a ring B. The shikimate pathway in plants leads to the biosynthesis of phenylpropanoids via the phenylpropanoid pathway. Phenylalanine, which comes from the shikimate pathway, acts as a precursor amino acid in the phenylpropanoid biosynthesis pathway, leading to the synthesis of flavonoids. This pathway is unique to plants because in addition to flavonoid biosynthesis, it also produces coumarins, esters, lignin, hydroxycinnamates, and other plant secondary metabolites [28]. Flavonoids are vital for plant survival and play an essential role in plant metabolism, especially in defense and pigmentation [29]. The phenylpropanoid production occurs in specialized plant tissues and cells; however, biosynthesis may also take place in response to environmental factors, such as ultraviolet (UV) damage or pathogenic attacks [30]. Naringenin chalcone also acts as a precursor molecule to produce flavonoids via the phenylpropanoid pathway [31]. The first step involved in the phenylpropanoid pathway is the phenylalanine ammonia-lyase (PAL)-mediated deamination of the amino acid phenylalanine to cinnamic acid. Next, cinnamate-4-hydroxylase (C4H) hydroxylates cinnamic acid to p-coumaric acid, which is later activated by para-coumarate-CoA ligase (PCL) to form p-coumaroyl-CoA. The step-by-step condensation of the malonyl-CoA three acetate units with p-coumaroyl-CoA is catalyzed by chalcone synthase (CHS), which ultimately results in the production of naringenin chalcone [32]. In the final step of flavonoid biosynthesis, chalcone isomerase (CHI) converts naringenin chalcone to flavanone or naringenin, as represented in Figure 2. Different flavonoids, such as flavones, isoflavones, and anthocyanidins, are further generated from various structural modifications of naringenin [33]. Due to the specific action of CHS, a variety of secondary metabolites are produced as part of the phenylpropanoid pathway [34,35][34][35]. The A and B phenyl rings, which form the backbone of the flavonoid structure, arise via the action of CHS, as it combines three molecules of malonyl-CoA with p-coumaroyl-CoA to form a chalcone [36]. CHI takes part in the synthesis of the important C ring of the flavonoid backbone resulting in the formation of flavanone (naringenin). Isoflavone synthase (IFS) catalyzes the transition of flavanone to isoflavone. Flavanone 3 β-hydroxylase (F3H) hydroxylates naringenin to form dihydroflavonol, which is then converted to leucoanthocyanidin by dihydroflavonol 4-reductase (DFR) [37]. Further catalysis mediated by anthocyanidin synthase (ANS) leads to the formation of anthocyanidin from leucoanthocyanidins, whereas leucoanthocyanidin reductase (LAR) undergoes the reduction of leucoanthocyanidins to flavan-3-ol (also referred to as flavanols, a complex class of flavonoids) [38].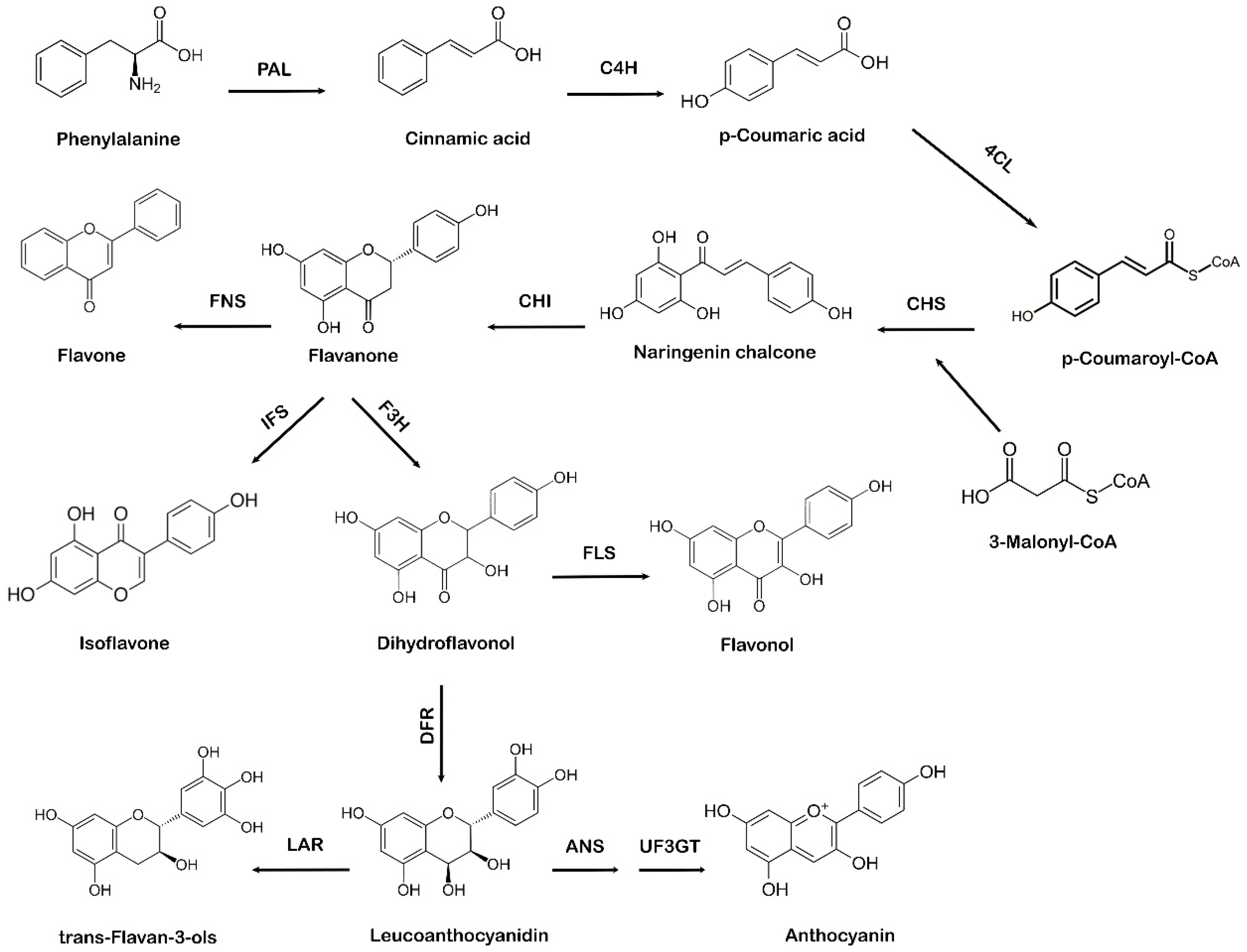
Figure 2. Systematic representation of flavonoid biosynthesis. PAL: phenylalanine ammonia-lyase, C4H: cinnamate-4-hydroxylase, 4CL: 4-coumarate:CoA ligase, CHS: chalcone synthase, CHI: chalcone isomerase, IFS: isoflavone synthase, F3H: flavanone 3 β-hydroxylase, FNS: flavone synthase, FLS: flavonol synthase, DFR: dihydroflavonol 4-reductase, ANS: anthocyanidin synthase, UF3GT: anthocyanidin 3-O-glucosyltransferase, LAR: leucoanthocyanidin reductase.
3. Microbial Co-Culture Strategy for Flavonoid Biosynthesis
Co-culture engineering offers a robust and efficient toolbox for complex compound biosynthesis with long biosynthetic pathways. The engineering of microbial consortia for the expression of complex biosynthetic pathways of flavonoids is a promising approach for flavonoid production and has been extensively researched [20,182][20][39]. However, existing microbial biosynthesis strategies depend primarily on the use of a single strain, generated via metabolic engineering, to accommodate the complicated and long flavonoid pathways, which is an efficient way to biosynthesize valuable compounds, but suffers from many drawbacks, such as severe metabolic imbalances, failure to achieve optimum conditions for all pathway-specific enzymes to function, increased metabolic burden because of the reconstruction or heterologous expression of complicated metabolic pathways, and unwanted by-product accumulation, making it difficult to increase the yields of the final product [128,182,183][39][40][41]. Recently, co-culture systems utilizing two or more microbial strains were developed to resolve these concerns and improve final titers [128][40] (Figure 3). Compared to the conventional monoculture strategy, modular co-culture engineering exploits the power of multiple strains of microbes to reconstruct the target biosynthetic pathway. As a result, it significantly reduces the biosynthesis labor of each microbial strain, as well as the metabolic burden associated with it, and it also restricts the formation of undesired by-products [183,184][41][42]. These benefits are exceptional for the biological synthesis of diverse natural plant products, involving intricate lengthy pathways, and requiring extensive yet delicate engineering efforts [182][39]. Microbial co-cultures provide numerous advantages over monocultures in terms of removing pathway bottlenecks and enhancing metabolic productivity. These advantages include metabolic labor division, the sharing of gene expression burdens, and the ability to allow cross-feeding of metabolites to help improve pathway performance and metabolic robustness. A complete biosynthetic pathway is modularized using the most extensively used co-culture approach, with each module tailored to achieve the maximum production volume and intermediate metabolite yield. In co-culture, metabolic intermediates or precursors must be available and transported to the downstream strains [125][43]. Moreover, cellular specialization and compartmentalization allow microbial consortia to cope with major environmental fluctuations and to perform complicated tasks that individual members would be unable to carry out. Thus, microbial co-cultures can improve productivity and precision by presenting a simple way of optimizing each submodule by dividing the metabolic burden and reducing the strain on members of the consortia present in the culture [125][43].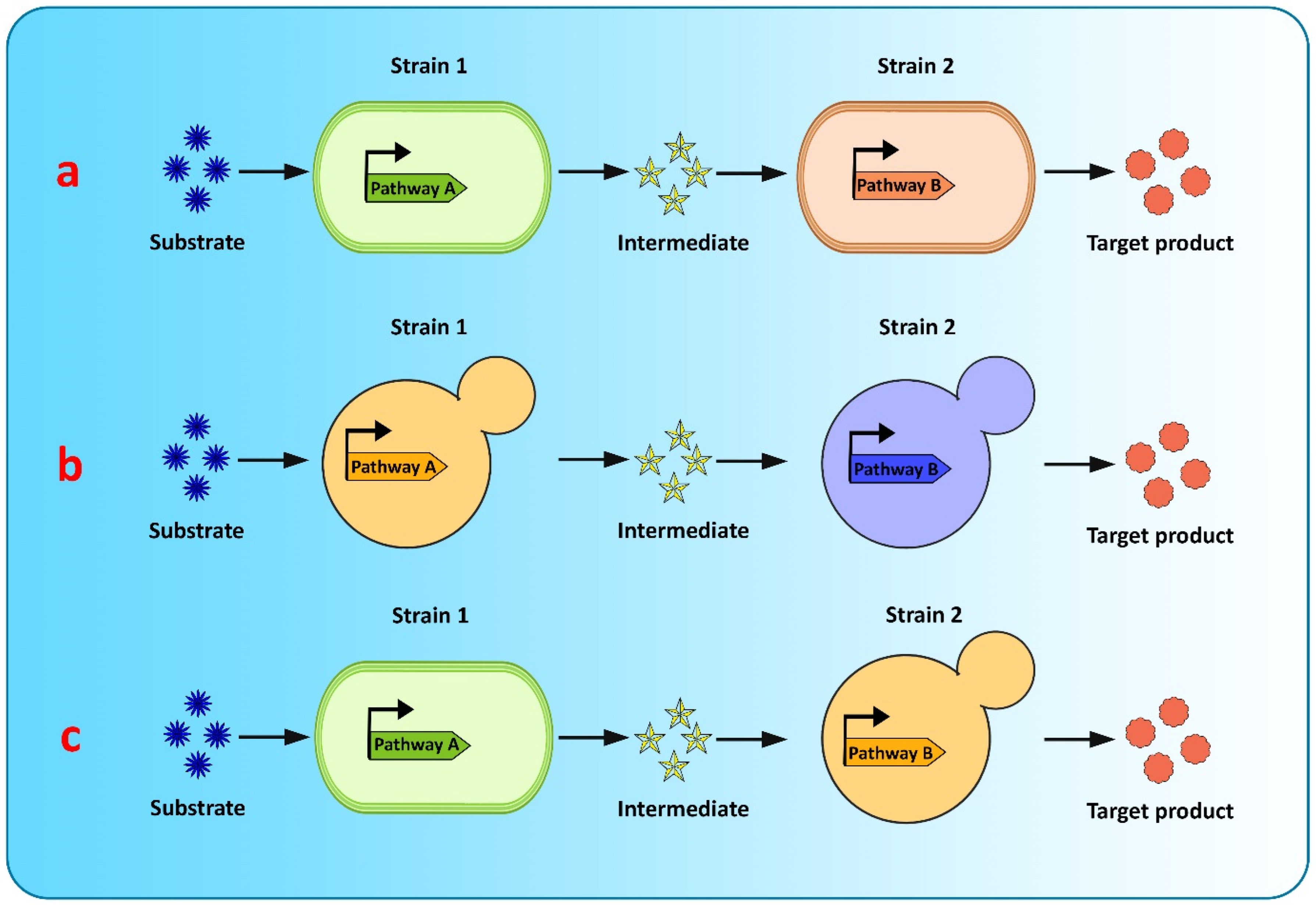
Figure 3. Graphical representation of modular co-culture approach for de novo biosynthesis of flavonoids (a) E. coli–E. coli co-culture (b) S. cerevisiae–S. cerevisiae co-culture (c) E. coli–S. cerevisiae co-culture.
Table 1. Co-culture approach for the microbial production of flavonoids.
| Co-Culture Strains | Substrate | Product | Titer (mg/L) | References |
|---|---|---|---|---|
| E. coli–E. coli coculture | Glucose | Sakuranetin | 29.7 | [182][39] |
| E. coli–E. coli coculture | (+)-Catechin and glucose | Pyranocyanidin-3-O-glucoside-catechol | 13 | [185][44] |
| E. coli–E. coli coculture | (+)-Catechin, glucose, and tyrosine | Pyranocyanidin-3-O-glucoside-phenol | 19.5 | [185][44] |
| E. coli–E. coli coculture | p-Coumaric acid | Apigetrin | 16.6 | [183][41] |
| E. coli–E. coli coculture | Apigenin and luteolin | Orientin | 7090 | [187][46] |
| E. coli–E. coli coculture | Apigenin and luteolin | Vitexin | 5050 | [187][46] |
| E. coli–S. cerevisiae coculture | Glucose | Icaritin | 19.7 | [180][47] |
| E. coli–S. cerevisiae coculture | Xylose | Naringenin | 21.16 | [188][48] |
| S. cerevisiae–S. cerevisiae coculture | Naringenin | Delphinidin | 26.1 | [20] |
| S. cerevisiae–S. cerevisiae coculture | p-Coumaric acid | Naringenin | 18.5 | [189][49] |
References
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as Anticancer Agents. Nutrients 2020, 12, 457.
- Borja, G.M.; Rodriguez, A.; Campbell, K.; Borodina, I.; Chen, Y.; Nielsen, J. Metabolic engineering and transcriptomic analysis of Saccharomyces cerevisiae producing p-coumaric acid from xylose. Microb. Cell Factories 2019, 18, 191.
- Caro-Ordieres, T.; Marín-Royo, G.; Opazo-Ríos, L.; Jiménez-Castilla, L.; Moreno, J.A.; Gómez-Guerrero, C.; Egido, J. The Coming Age of Flavonoids in the Treatment of Diabetic Complications. J. Clin. Med. 2020, 9, 346.
- Gujar, K.; Wairkar, S. Nanocrystal technology for improving therapeutic efficacy of flavonoids. Phytomedicine 2020, 71, 153240.
- Shah, F.L.A.; Ramzi, A.B.; Baharum, S.N.; Noor, N.M.; Goh, H.-H.; Leow, T.C.; Oslan, S.N.; Sabri, S. Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol. Biol. Rep. 2019, 46, 6647–6659.
- Ciumărnean, L.; Milaciu, M.V.; Runcan, O.; Vesa, Ș.C.; Răchișan, A.L.; Negrean, V.; Perné, M.-G.; Donca, V.I.; Alexescu, T.-G.; Para, I.; et al. The Effects of Flavonoids in Cardiovascular Diseases. Molecules 2020, 25, 4320.
- Pei, R.; Liu, X.; Bolling, B. Flavonoids and gut health. Curr. Opin. Biotechnol. 2020, 61, 153–159.
- Al Aboody, M.S.; Mickymaray, S. Anti-Fungal Efficacy and Mechanisms of Flavonoids. Antibiotics 2020, 9, 45.
- Boojar, M.M.A. An overview of the cellular mechanisms of flavonoids radioprotective effects. Adv. Pharm. Bull. 2020, 10, 13.
- Alexey, D.; Paula, G.; Ana Rute, N.; Jochen, F. Engineering of Microbial Cell Factories for the Production of Plant Polyphenols with Health-Beneficial Properties. Curr. Pharm. Des. 2018, 24, 2208–2225.
- Marsafari, M.; Samizadeh, H.; Rabiei, B.; Mehrabi, A.; Koffas, M.; Xu, P. Biotechnological Production of Flavonoids: An Update on Plant Metabolic Engineering, Microbial Host Selection, and Genetically Encoded Biosensors. Biotechnol. J. 2020, 15, 1900432.
- De Bruijn, W.J.C.; Levisson, M.; Beekwilder, J.; van Berkel, W.J.H.; Vincken, J.-P. Plant Aromatic Prenyltransferases: Tools for Microbial Cell Factories. Trends Biotechnol. 2020, 38, 917–934.
- Uddin, M.S.; Kabir, M.T.; Niaz, K.; Jeandet, P.; Clément, C.; Mathew, B.; Rauf, A.; Rengasamy, K.R.R.; Sobarzo-Sánchez, E.; Ashraf, G.M.; et al. Molecular Insight into the Therapeutic Promise of Flavonoids against Alzheimer’s Disease. Molecules 2020, 25, 1267.
- Dan, W.; Dai, J. Recent developments of chalcones as potential antibacterial agents in medicinal chemistry. Eur. J. Med. Chem. 2020, 187, 111980.
- Ninfali, P.; Antonelli, A.; Magnani, M.; Scarpa, E.S. Antiviral Properties of Flavonoids and Delivery Strategies. Nutrients 2020, 12, 2534.
- Musumeci, L.; Maugeri, A.; Cirmi, S.; Lombardo, G.E.; Russo, C.; Gangemi, S.; Calapai, G.; Navarra, M. Citrus fruits and their flavonoids in inflammatory bowel disease: An overview. Nat. Prod. Res. 2020, 34, 122–136.
- Spagnuolo, C.; Moccia, S.; Russo, G.L. Anti-inflammatory effects of flavonoids in neurodegenerative disorders. Eur. J. Med. Chem. 2018, 153, 105–115.
- Mark, R.; Lyu, X.; Ng, K.R.; Chen, W.N. Gene Source Screening as a Tool for Naringenin Production in Engineered Saccharomyces cerevisiae. ACS Omega 2019, 4, 12872–12879.
- Tian, B.; Pei, Y.; Huang, W.; Ding, J.; Siemann, E. Increasing flavonoid concentrations in root exudates enhance associations between arbuscular mycorrhizal fungi and an invasive plant. ISME J. 2021, 15, 1919–1930.
- Du, Y.; Yang, B.; Yi, Z.; Hu, L.; Li, M. Engineering Saccharomyces cerevisiae Coculture Platform for the Production of Flavonoids. J. Agric. Food Chem. 2020, 68, 2146–2154.
- Singh, B.; Kumar, A.; Malik, A.K. Flavonoids biosynthesis in plants and its further analysis by capillary electrophoresis. Electrophoresis 2017, 38, 820–832.
- Sajid, M.; Channakesavula, C.N.; Stone, S.R.; Kaur, P. Synthetic biology towards improved flavonoid pharmacokinetics. Biomolecules 2021, 11, 754.
- Bahaudin, K.; Sabri, S.; Ramzi, A.B.; Chor, A.L.T.; Tencomnao, T.; Baharum, S.N. Current progress in production of flavonoids using systems and synthetic biology platforms. Sains Malays. 2018, 47, 3077–3084.
- Muhammad, A.; Feng, X.; Rasool, A.; Sun, W.; Li, C. Production of plant natural products through engineered Yarrowia lipolytica. Biotechnol. Adv. 2020, 43, 107555.
- Cao, M.; Gao, M.; Suástegui, M.; Mei, Y.; Shao, Z. Building microbial factories for the production of aromatic amino acid pathway derivatives: From commodity chemicals to plant-sourced natural products. Metab. Eng. 2020, 58, 94–132.
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The Flavonoid Biosynthesis Network in Plants. Int. J. Mol. Sci. 2021, 22, 12824.
- Wang, X.; Ding, G.; Liu, B.; Wang, Q. Flavonoids and antioxidant activity of rare and endangered fern: Isoetes sinensis. PLoS ONE 2020, 15, e0232185.
- Venkateswara Rao, P.; Kiran, S.; Rohini, P.; Bhagyasree, P. Flavonoid: A review on Naringenin. J. Pharmacogn. Phytochem. 2017, 6, 2778–2783.
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316.
- Mathesius, U. Flavonoid Functions in Plants and Their Interactions with Other Organisms. Plants 2018, 7, 30.
- Ibdah, M.; Martens, S.; Gang, D.R. Biosynthetic Pathway and Metabolic Engineering of Plant Dihydrochalcones. J. Agric. Food Chem. 2018, 66, 2273–2280.
- Deng, Y.; Li, C.; Li, H.; Lu, S. Identification and Characterization of Flavonoid Biosynthetic Enzyme Genes in Salvia miltiorrhiza (Lamiaceae). Molecules 2018, 23, 1467.
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028.
- Trantas, E.A.; Koffas, M.A.G.; Xu, P.; Ververidis, F. When plants produce not enough or at all: Metabolic engineering of flavonoids in microbial hosts. Front. Plant Sci. 2015, 6, 7.
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47.
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The Origin and Evolution of Plant Flavonoid Metabolism. Front. Plant Sci. 2019, 10, 943.
- Teles, Y.C.F.; Souza, M.S.R.; Souza, M.D.F.V.D. Sulphated Flavonoids: Biosynthesis, Structures, and Biological Activities. Molecules 2018, 23, 480.
- Rana, A.C.; Gulliya, B. Chemistry and pharmacology of flavonoids—A review. Indian J. Pharm. Educ. Res. 2019, 53, 8–20.
- Wang, X.; Li, Z.; Policarpio, L.; Koffas, M.A.G.; Zhang, H. De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl. Microbiol. Biotechnol. 2020, 104, 4849–4861.
- Kim, B.-G. Biological synthesis of genistein in Escherichia coli. J. Microbiol. Biotechnol. 2020, 30, 770–776.
- Thuan, N.H.; Chaudhary, A.K.; Van Cuong, D.; Cuong, N.X. Engineering co-culture system for production of apigetrin in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2018, 45, 175–185.
- Wu, G.; Yan, Q.; Jones, J.A.; Tang, Y.J.; Fong, S.S.; Koffas, M.A.G. Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications. Trends Biotechnol. 2016, 34, 652–664.
- Xu, P.; Marsafari, M.; Zha, J.; Koffas, M. Microbial Coculture for Flavonoid Synthesis. Trends Biotechnol. 2020, 38, 686–688.
- Akdemir, H.; Silva, A.; Zha, J.; Zagorevski, D.V.; Koffas, M.A.G. Production of pyranoanthocyanins using Escherichia coli co-cultures. Metab. Eng. 2019, 55, 290–298.
- Thuan, N.H.; Tatipamula, V.B.; Viet, T.T.; Tien, N.Q.D.; Loc, N.H. Bioproduction of eriodictyol by Escherichia coli engineered co-culture. World J. Microbiol. Biotechnol. 2022, 38, 112.
- Qiu, C.; Wang, H.; Zhao, L.; Pei, J. Orientin and vitexin production by a one-pot enzymatic cascade of a glycosyltransferase and sucrose synthase. Bioorg. Chem. 2021, 112, 104926.
- Wang, P.; Li, C.; Li, X.; Huang, W.; Wang, Y.; Wang, J.; Zhang, Y.; Yang, X.; Yan, X.; Wang, Y. Complete biosynthesis of the potential medicine icaritin by engineered Saccharomyces cerevisiae and Escherichia coli. Sci. Bull. 2021, 66, 1906–1916.
- Zhang, W.; Liu, H.; Li, X.; Liu, D.; Dong, X.T.; Li, F.F.; Wang, E.X.; Li, B.Z.; Yuan, Y.J. Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng. Life Sci. 2017, 17, 1021–1029.
- Lu, Y.; Song, Y.; Zhu, J.; Xu, X.; Pang, B.; Jin, H.; Jiang, C.; Liu, Y.; Shi, J. Potential application of CHS and 4CL genes from grape endophytic fungus in production of naringenin and resveratrol and the improvement of polyphenol profiles and flavour of wine. Food Chem. 2021, 347, 128972.
More
