Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 3 by Conner Chen.
Methylmercury (MeHg) is an environmental neurotoxin that can adversely affect the development of the nervous system. The molecular integrity of chromatin in the nucleus is an important target of MeHg. Low levels of MeHg trigger epigenetic mechanisms that may be involved in long-lasting and transgenerational neurotoxicity after exposure. Emerging evidence has shown that these mechanisms include histone modification, short interfering RNA (siRNA)RNA, and DNA methylation.
- methylmercury
- neurotoxicity
- transgenerational
1. Introduction
Methylmercury (MeHg) is an environmental neurotoxin that may cause long-lasting neurotoxicity in vulnerable populations, such as childbearing women and children [1]. MeHg is a persistent toxicant that is produced from microorganisms in the water system [2]. Environmental exposure to MeHg in the general population comes from eating fish that contain various levels of MeHg (0.02~0.55 ppm; US EPA guideline: 0.30 ppm) [3]. The developing nervous system is a preferential target of MeHg-induced toxicity [4]. The toxic mechanisms of MeHg involves the attachment of MeHg to thiol groups in biomolecules, forming various MeHg-SR complexes [2]. MeHg-SR complexes have lipophilic properties, which tend to partition into cellular lipid regions [5]. Indeed, the increase in partition coefficient after binding (even to cysteine) can explain the LAT1-independent entrance of MeHg in cells [5]. Exposure to this organic mercurial results in the distribution of the metal in the lysosome, mitochondria, and nucleus [6][7][8]. The mechanisms of MeHg-induced cytotoxicity are related to the disruption of homeostasis in a host of cellular physiological functions, including glutathione (GSH) depletion [9], calcium overload [10], loss of mitochondria membrane potential [11][12], and endoplasmic reticulum (ER) stress [13][14]. MeHg can readily cross the nuclear envelope to bind with chromatin components [15][16]. The process may not be involved in the acute toxicity of the metal; however, it could induce changes in the structure of chromatin, leading to long-lasting effects after exposure [17][18][19].
2. MeHg, DNA, and Chromatin
MeHg has a characteristic binding activity to the C- or N-containing moiety groups in the bases of DNA [20]. The physicochemical property of MeHg makes it very useful in the study of DNA structure. One of its compounds, methylmercury hydroxylate (MeHgOH), was used as a chemical probe to investigate DNA secondary structure and unpaired bases [21]. As it readily reacts with the purine and pyrimidine residues of nucleic acids, MeHgOH was also used as a reversible denaturing agent for DNA agarose gel electrophoresis [22]. The binding of MeHgOH to different bases in singular and duplex DNA was utilized for the detection and quantification of single-stranded DNA in duplex DNA samples [16]. These in vitro binding studies, which employed MeHgOH as the chemical species of MeHg, are in line with the genotoxic effects of MeHg [23][24][25][26][27]. The binding property to DNA can be changed with different ligands complexed to MeHg: for example, methylmercury chloride (MeHgCl), another widely used experimental chemical species. MeHgCl can interact with cysteine to form a major bioavailable form, MeHg-S-Cys [28]. The exchange of dissociable anions in MeHg complexes with thiol groups in other biomolecules dictates its molecular toxicity [2][29]. A cell culture study has shown that both MeHgOH and MeHgCl can cause cytotoxicity and genotoxicity, and MeHgCl is more toxic to SH-SY5Y cells than MeHgOH [30]. The differential toxic effects between MeHgOH and MeHgCl can be attributed to the stronger lipid partition coefficient of MeHgCl [5]; however, in a complex system in the cell, the genotoxic effects are probably mediated by their interaction with thiol-containing critical chromatin proteins (Figure 1) [2]. The integrity of DNA replication may be compromised after MeHg exposure, as corroborated in many studies on MeHg-induced genotoxicity [23][24][25][26][27]. These studies pave the way for the understanding of MeHg-induced genotoxic effects and its potential impact on the structure of nucleosomes and chromatin.
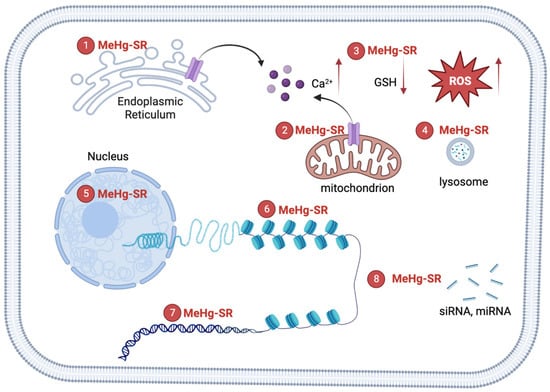
Figure 1. Mechanisms of MeHg-induced neurotoxicity. The formation of MeHg-SR complexes with endogenous thiol-containing biomolecules may increase its lipophilicity, resulting the distribution of the metal into hydrophobic compartments including mitochondria, lysosome, nucleus and other organelles. ① The complexation of MeHg with nascent proteins in the ER may cause ER stress. The black arrow (left): the flow of calcium ions from ER. ② MeHg-SR disrupts mitochondrial respiratory apparatus, leading to the elevation of reactive oxygen species (ROS). Multiples sources of Ca2+ contribute to MeHg-induced increase in intracellular Ca2+ [10]. The black arrow (right): the flow of calcium ions from mitochondrion. The red arrows (left): the increase of calcium ions; (middle): the decrease of GSH; (right): the increase of ROS. ③ The exchange of MeHg-SR with glutathione (GSH) results in reduction of GSH levels [9]. ④ The majority of MeHg-SR resides in lysosome, which corroborates the structural damage to biomolecules through attachment of the metal to thiol groups [8]. ⑤ MeHg-SR in the nucleus has the potential effects on the integrity of chromatin structures by complexing with histones or DNA [15][31][32]. ⑥ MeHg-RS changes histone post-translational modifications to affect the compactness of chromatin [15]. ⑦ A direct binging MeHg to bases of DNA constitutes the molecular basis for genotoxicity [20]. ⑧ MeHg-SR can interrupt biogenesis of short interfering RNA (siRNA)RNA and/or miRNA, leading to alterations in siRNA and/or miRNA-mediated gene regulations [33].
Although MeHg has a high affinity for sulfhydryl groups [34], its binding partners in the cellular system are also governed by a number of factors, such as exchange reactions and protein-specific structural and thermodynamical factors [2]. MeHg can bind with a variety of biomolecules in the nucleus, including DNA, histones, and non-histone protein components [15][31][35]. Likewise, many factors influence the propensity of the binding of MeHg to DNA bases, including the concentration of MeHg, temperature, base composition, ionic strength, and pH, to name a few [20]. A study in HeLa S3 suspension-culture cells has shown that the binding of MeHgCl to DNA and chromatin is a rapid process and could readily take place after the exposure [31]. A primary cell culture study in mouse fetal astrocyte has shown that MeHgCl exposure can competitively block the histone binding sites of the histone specific dye, N-(3-pyrene)maleimide, which specifically labels the cysteine groups in histone H3 of nucleosomes [15]. The blockage of the dye binding was gradually increased upon prolonged exposures in the in vitro model [15]. MeHg exposure also disrupts the complexing of histones with DNA. An in vitro study has shown that MeHgOH (1~10 μM) interferes with the binding of DNA by histones H3 and H4 [32]. A higher concentration of MeHgOH (10~32 μM) disrupted the complexing of DNA with the histones H2A and H2B [32]. The structure of chromatin plays an important role in gene regulation, which is predominantly regulated by histone proteins [36]. The post-translational modifications of histones, such as methylation and acetylation, regulates chromatin compactness, transcriptional activity, and genome functions [37]. Further, the regulation of gene expression at the transcriptional level involves epigenetic programs that are encoded by histone post-translational modifications, which determine the compactness of chromatin and transcriptional activity [38]. For example, increases in histone H3K4 methylation around the transcriptional start site (TSS) are associated with active transcription while high levels of H3K9 methylation at this region are associated with gene repression [39]. The regulatory mechanism during neuronal development and differentiation spatially and temporally invokes the modification of histones to fulfill gene regulation purposes [40]. Thus, the potential effects of MeHg on the post-translational modification of histones likely involves a direct binding of MeHg to the components of chromatin (particularly to thiol-containing proteins), leading to interference in chromatin compactness and structure. Though MeHg can interact with nucleophilic N-atoms found in nitrogenous bases forming the DNA nucleotides, the thiol-containing proteins found in the chromatin are possibly MeHg’s preferential targets. However, the knowledge on how MeHg disrupts the physiology and biochemistry of chromatin is still elusive. Recent experimental studies have shown that low levels of MeHg (nM) induce changes in histone post-translational modifications and DNA methylation, both of which may serve an important base for the long-lasting and transgenerational effects after the exposure [17][18].
References
- Cediel Ulloa, A.; Gliga, A.; Love, T.M.; Pineda, D.; Mruzek, D.W.; Watson, G.E.; Davidson, P.W.; Shamlaye, C.F.; Strain, J.J.; Myers, G.J.; et al. Prenatal methylmercury exposure and DNA methylation in seven-year-old children in the Seychelles Child Development Study. Environ. Int. 2021, 147, 106321.
- Nogara, P.A.; Oliveira, C.S.; Schmitz, G.L.; Piquini, P.C.; Farina, M.; Aschner, M.; Rocha, J.B.T. Methylmercury’s chemistry: From the environment to the mammalian brain. Biochim. Biophys. Acta BBA Gen. Subj. 2019, 1863, 129284.
- Barone, G.; Storelli, A.; Meleleo, D.; Dambrosio, A.; Garofalo, R.; Busco, A.; Storelli, M.M. Levels of Mercury, Methylmercury and Selenium in Fish: Insights into Children Food Safety. Toxics 2021, 9, 39.
- Ortega-García, J.A.; Rodriguez, K.; Calatayud, M.; Martin, M.; Vélez, D.; Devesa, V.; Sánchez-Alarcon, M.C.; Torres Cantero, A.M.; Galindo-Cascales, C.; Gil-Vázquez, J.M.; et al. Estimated intake levels of methylmercury in children, childbearing age and pregnant women in a Mediterranean region, Murcia, Spain. Eur. J. Pediatr. 2009, 168, 1075–1080.
- Mason, R.P. The Bioaccumulation of Mercury, Methylmercury, and Other Toxic Elements into Pelagic and Benthic Organisms. In Coastal and Estuarine Risk Assessment; CRC Press: Boca Raton, FL, USA, 2001; pp. 143–166.
- Raes, B.B. The ultrastructural effect and subcellular localization of mercuric chloride and methylmercuric chloride in insect cells (Aedes albopictus C6/36). Tissue Cell 1999, 31, 223–232.
- Komsta-Szumska, E.; Reuhl, K.R.; Miller, D.R. The effect of methylmercury on the distribution and excretion of selenium by the guinea pig. Arch. Toxicol. 1983, 54, 303–310.
- Eto, K. Pathology of Minamata disease. Toxicol. Pathol. 1997, 25, 614–623.
- Farina, M.; Aschner, M. Glutathione antioxidant system and methylmercury-induced neurotoxicity: An intriguing interplay. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 129285.
- Yuan, Y.; Atchison, W.D. Multiple Sources of Ca2+ Contribute to Methylmercury-Induced Increased Frequency of Spontaneous Inhibitory Synaptic Responses in Cerebellar Slices of Rat. Toxicol. Sci. 2016, 150, 117–130.
- Lee, J.Y.; Ishida, Y.; Takahashi, T.; Naganuma, A.; Hwang, G.W. Transport of pyruvate into mitochondria is involved in methylmercury toxicity. Sci. Rep. 2016, 6, 21528.
- Ke, T.; Rocha, J.B.T.; Tinkov, A.A.; Santamaria, A.; Bowman, A.B.; Aschner, M. The Role of Human LRRK2 in Acute Methylmercury Toxicity in Caenorhabditis elegans. Neurochem. Res. 2021, 46, 2991–3002.
- Usuki, F.; Fujimura, M.; Yamashita, A. Endoplasmic reticulum stress preconditioning modifies intracellular mercury content by upregulating membrane transporters. Sci. Rep. 2017, 7, 12390.
- Takanezawa, Y.; Nakamura, R.; Hamaguchi, M.; Yamamoto, K.; Sone, Y.; Uraguchi, S.; Kiyono, M. Docosahexaenoic acid enhances methylmercury-induced endoplasmic reticulum stress and cell death and eicosapentaenoic acid potentially attenuates these effects in mouse embryonic fibroblasts. Toxicol. Lett. 2019, 306, 35–42.
- Choi, B.H.; Simpkins, H. Changes in the molecular structure of mouse fetal astrocyte nucleosomes produced in vitro by methylmercuric chloride. Env. Res. 1986, 39, 321–330.
- Maki, A.H.; Ott, C.M. Methylmercury(II) binding to single-stranded and duplex DNA: Complexes formed are distinguishable by optical detection of magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA 1981, 78, 2972–2976.
- Carvan, M.J., 3rd; Kalluvila, T.A.; Klingler, R.H.; Larson, J.K.; Pickens, M.; Mora-Zamorano, F.X.; Connaughton, V.P.; Sadler-Riggleman, I.; Beck, D.; Skinner, M.K. Mercury-induced epigenetic transgenerational inheritance of abnormal neurobehavior is correlated with sperm epimutations in zebrafish. PLoS ONE 2017, 12, e0176155.
- Onishchenko, N.; Karpova, N.; Sabri, F.; Castrén, E.; Ceccatelli, S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J. Neurochem. 2008, 106, 1378–1387.
- Bose, R.; Onishchenko, N.; Edoff, K.; Janson Lang, A.M.; Ceccatelli, S. Inherited effects of low-dose exposure to methylmercury in neural stem cells. Toxicol. Sci. 2012, 130, 383–390.
- Onyido, I.; Norris, A.R.; Buncel, E. Biomolecule-mercury interactions: Modalities of DNA base-mercury binding mechanisms. Remediation strategies. Chem. Rev. 2004, 104, 5911–5929.
- Beerman, T.A.; Lebowitz, J. Further analysis of the altered secondary structure of superhelical DNA. Sensitivity to methylmercuric hydroxide a chemical probe for unpaired bases. J. Mol. Biol. 1973, 79, 451–470.
- Bailey, J.M.; Davidson, N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal. Biochem. 1976, 70, 75–85.
- Vicari, T.; Ferraro, M.V.; Ramsdorf, W.A.; Mela, M.; de Oliveira Ribeiro, C.A.; Cestari, M.M. Genotoxic evaluation of different doses of methylmercury (CH₃Hg⁺) in Hoplias malabaricus. Ecotoxicol. Environ. Saf. 2012, 82, 47–55.
- Ondovcik, S.L.; Tamblyn, L.; McPherson, J.P.; Wells, P.G. Oxoguanine glycosylase 1 (OGG1) protects cells from DNA double-strand break damage following methylmercury (MeHg) exposure. Toxicol. Sci. 2012, 128, 272–283.
- Lerebours, A.; Cambier, S.; Hislop, L.; Adam-Guillermin, C.; Bourdineaud, J.P. Genotoxic effects of exposure to waterborne uranium, dietary methylmercury and hyperoxia in zebrafish assessed by the quantitative RAPD-PCR method. Mutat. Res. 2013, 755, 55–60.
- Crespo-Lopez, M.E.; Costa-Malaquias, A.; Oliveira, E.H.; Miranda, M.S.; Arrifano, G.P.; Souza-Monteiro, J.R.; Sagica, F.E.; Fontes-Junior, E.A.; Maia, C.S.; Macchi, B.M.; et al. Is Low Non-Lethal Concentration of Methylmercury Really Safe? A Report on Genotoxicity with Delayed Cell Proliferation. PLoS ONE 2016, 11, e0162822.
- Sousa, A.H.; Pereira, J.P.G.; Malaquias, A.C.; Sagica, F.; de Oliveira, E.H.C. Intracellular accumulation and DNA damage caused by methylmercury in glial cells. J. Biochem. Mol. Toxicol. 2022, 36, e23170.
- Yin, Z.; Jiang, H.; Syversen, T.; Rocha, J.B.; Farina, M.; Aschner, M. The methylmercury-L-cysteine conjugate is a substrate for the L-type large neutral amino acid transporter. J. Neurochem. 2008, 107, 1083–1090.
- Rabenstein, D.L. Chemistry of methylmercury toxicology. J. Chem. Educ. 1978, 55, 292.
- Patnaik, R.; Padhy, R.N. Comparative study on toxicity of methylmercury chloride and methylmercury hydroxide to the human neuroblastoma cell line SH-SY5Y. Environ. Sci. Pollut. Res. Int. 2018, 25, 20606–20614.
- Gruenwedel, D.W.; Glaser, J.F.; Cruikshank, M.K. Binding of methylmercury(II) by HeLa S3 suspension-culture cells: Intracellular methylmercury levels and their effect on DNA replication and protein synthesis. Chem. Biol. Interact. 1981, 36, 259–274.
- Otsuki, L.G.; Gruenwedel, D.W. Methylmercury-chromosome interactions. I. Thermal denaturation of calf thymus chromatin in presence of CH3HgOH. Z Nat. C Biosci. 1980, 35, 605–610.
- Rudgalvyte, M.; VanDuyn, N.; Aarnio, V.; Heikkinen, L.; Peltonen, J.; Lakso, M.; Nass, R.; Wong, G. Methylmercury exposure increases lipocalin related (lpr) and decreases activated in blocked unfolded protein response (abu) genes and specific miRNAs in Caenorhabditis elegans. Toxicol. Lett. 2013, 222, 189–196.
- Lemes, M.; Wang, F. Methylmercury speciation in fish muscle by HPLC-ICP-MS following enzymatic hydrolysis. J. Anal. At. Spectrom. 2009, 24, 663–668.
- Chanda, S.K.; Cherian, M.G. Isolation and partial characterization of a mercury-binding nonhistone protein component from rat kidney nuclei. Biochem. Biophys. Res. Commun. 1973, 50, 1013–1019.
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189.
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580.
- Weber, C.M.; Henikoff, S. Histone variants: Dynamic punctuation in transcription. Genes Dev. 2014, 28, 672–682.
- Lim, P.S.; Shannon, M.F.; Hardy, K. Epigenetic control of inducible gene expression in the immune system. Epigenomics 2010, 2, 775–795.
- Park, J.; Lee, K.; Kim, K.; Yi, S.-J. The role of histone modifications: From neurodevelopment to neurodiseases. Signal Transduct. Target. Ther. 2022, 7, 217.
More
