γTubulin is part of a family of GTPases called the tubulins. Due to the self-polymerizing ability of γ-tubulin, in combination with its presence in all cellular compartments, γtubulin can be organized in γ-strings, and γ-tubules, and the γ-strings associated with the centrosome. These components are interlinked to form a cellular meshwork in both the cytosol (including all cellular organelles) and the nuclear compartment [14,17,18].
- γ-tubulin
- nuclear architecture
- cytoskeleton
- nuclearskeleton
- cancer
1. Definition
γTubulin is part of a family of GTPases called the tubulins, which are involved in shaping the architecture of the human centrosome [1][2][81,82]. In humans, there are five known tubulin isoforms, αtubulin, βtubulin, γtubulin, δtubulin, and εtubulin [3][83], of which only α-, β-, and γ-tubulins are ubiquitous. Multiple genes encode for α- and for β-tubulin, but the number of genes encoding γtubulin ranges from one to three, with two genes found in mammals and up to three genes found in flowering plants (http://genome.ucsc.edu/) [4][84].
Due to the self-polymerizing ability of γ-tubulin, in combination with its presence in all cellular compartments, γtubulin can be organized in γ-strings, and γ-tubules, and the γ-strings associated with the centrosome. These components are interlinked to form a cellular meshwork in both the cytosol (including all cellular organelles) and the nuclear compartment [5][6][7][14,17,18].
2. Introduction
The compartmentalization of eukaryotic cells into several organelles creates the need for structure, communication, and transport between compartments. In the cytoplasm and nucleus, three main families of structures ensure these functions—actin filaments, intermediate filaments, and MTs—and the LINCs are one of the linking structures between compartments [8][80]. In addition, the γtubulin meshwork establishes a connection between the cytoplasmic and nuclear compartments [5][6][7] [14,17,18].
Studies of human U2OS osteosarcoma cells and murine NIH3T3 embryonic fibroblasts revealed that in the tubulin family, γtubulin is the only member that contains an NLS [9][85] and a helix-loop-helix DNA-binding motif on the C terminus [10][86]. In humans, two proteins, γtubulin1 and γtubulin2, are encoded by two genes: TUBG1 and TUBG2. γTubulin1 is a ubiquitously expressed protein, whereas γtubulin2 is highly expressed in the brain [11][87]. γTubulin1 and γtubulin2 proteins exhibit an amino acid level identity of 97%, and the main differences in the protein sequence are localized to the DNA binding domain at the C-terminus of the protein [5][12][14,88].
The shared characteristics among αtubulin, βtubulin, and γtubulin explain the similarities between MTs and the γtubulin meshwork. In eukaryotic cells, α- and β-tubulin heterodimers and monomers of γtubulins form protofilaments [11][13][14][15][87,89,90,91] and γstrings, respectively (Figure 1). γStrings are located in the cytoplasm, the PeriCentriolar Material (PCM) of the centrosome, and the nuclear compartment (Figure 1 and Figure 2 and Table 1) [5][6][7][14,17,18], whereas cytosolic α- and β-tubulin protofilaments nucleate on a γTuRC [11][13][14][15][87,89,90,91] to form a MT (Table 1) [16][17][18][92,93,94]. In mammalian cells, in the absence of α- and β-tubulin heterodimers, γTuRCs and pericentrin assemble a γtubule (Figure 2), resulting in the formation of a fiber of similar size as an MT [19][16]. γTuRCs are found in the cytoplasm and the centrosomes, and are also associated with cellular membranes [11][13][14][15][87,89,90,91]. In contrast to γstrings, which are static structures, γtubules and MTs are temperature-sensitive polar structures that vary both in size and location, and both can emanate from centrosomes (Figure 2) [19][20][16,95].
Table 1. Proteins and protein complexes that both interact with γtubulin and affect nuclear structure.
| Protein/Complex | Reference | Comments |
|---|
| Centrosome | [2][21] | [82,83] | Cytoskeletal organizing centers |
| Microtubules | [16][17][18] | [92,93,94] | Polymers of tubulins |
| γTuRC | [11][13][14][15] | [87,89,90,91] | MT-nucleating unit |
| γTubule | [19] | [16] | γTubulin-rich filaments |
| γStrings | [6][7][22] | [17,18,96] | Thin cytosolic/nuclear γtubulin threads |
| Actin | [23] | [97] | Cytoskeletal/nuclearskeletal element |
| Intermediate filaments | [24] | [98] | Cytoskeleton |
| Lamin B | [6] | [17] | Nuclearskeleton |
| Ran | [25] | [99] | Regulates transport across NPC |
| Mel-28/ELYS | [25] | [99] | Required for NPC assembly |
| Nucleolin | [26][27] | [100,101] | RNA-binding protein at the nucleolus |
| E2F | [12][28][29][30][31] | [88,102,103] | Regulates gene expression |
| SUN | [32] | [33] | Links the nuclear lamina with NPCs |
| Samp1 | [30][31] | [104,105] | Inner nuclear membrane protein |
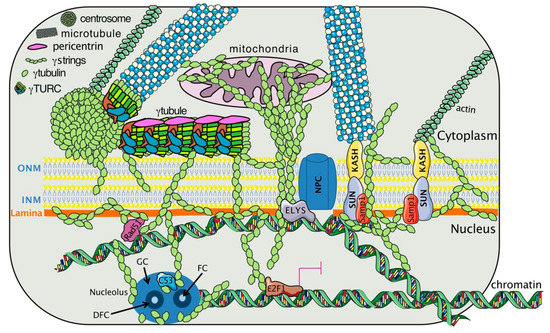
Figure 1. The γtubulin meshwork interacts with cellular components that affect the nuclear architecture. The meshwork is composed of centrosomes, γtubules, and γstrings. A γtubule consists of a γtubulin ring complex (γTURC), and pericentrin and can lie close to the outer nuclear membrane (ONM). Hypothetical representation of how γstrings connect cytosolic organelles (centrosome, mitochondria) and cytoskeletal elements (MTs, actin, and γtubules) with the nuclear compartment [5][6][7][14,17,18]. In the nucleus, γtubulin interacts with laminB (lamina) [33][17], the nucleoporin embryonic large molecule derived from yolk sac (ELYS), which is part of the nuclear pore complex (NPC) [34][99], the inner nuclear membrane (INM) protein Samp1 [35][104], the chromatin associated proteins Rad51 [36] [106] and C53 [37][100], and the transcription factor E2 promoter binding factor (E2F)1 [12][88]. The black arrows indicate the positions of the fibrillar center (FC), granular center (GC), and dense fibrillar component (DFC) in the nucleolus. The magenta lines indicate inhibition.
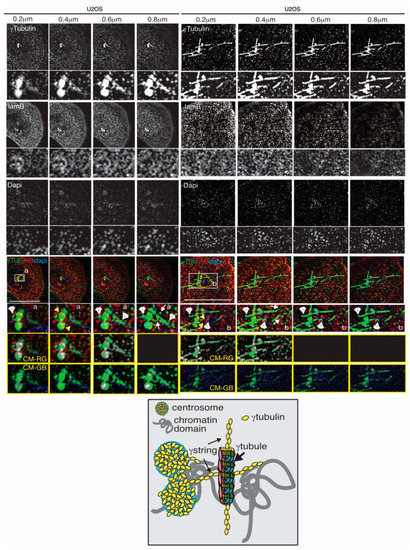
Figure 2. Chromosomes, centrosomes, γtubules, and γstrings are connected and facilitate the formation of chromatin domains (yellow dashed line). Structured illumination microscopy images of two fixed U2OS cells show the localization of endogenous γtubulin with an anti-γtubulin (γTub), lamina with the anti-laminB (lmB; a marker of the nuclear membrane) antibody, and DNA with DAPI in U2OS cells in the interphase. Sequential confocal images (6 stacks per cell) were collected at 0.2 μm intervals to enable a comparison of the network of fibers. White arrowheads and arrows show γtubules and γstrings, respectively, and yellow arrows indicate the centrosomes. White boxes show the magnified areas displayed in the insets. The scale bar is 10 μm. The yellow boxes show colocalized pixel maps (CM) of the red (R), green (G), and blue (B) channels of the magnified areas illustrated in the inset. Grey areas in the maps denote pixels colocalized between channels. The cartoon represents interactions between γtubules, γstrings, and centrosomes at the NE, which act as a protein scaffold for the formation of chromatin domains.
3. The Dynamics of the Gamma-Tubulin Meshwork
Cellular γtubulin has been described as being associated with all of the following compartments: the nucleus, the Golgi, the endoplasmic reticulum, the endosomes, the mitochondria, and the centrosomes (Table 1) [7][38][13][14][39][27][35][36][40][18,88,89,90,91,101,104,106,107]. Due to its self-polymerizing features, γtubulin produced by bacteria assembles in vitro γstrings that support the formation of lamin B3 protofilaments (Table 1) [6][17].
During cell division, the inherited centrosome and genome duplicate synchronously in the S-phase. At the onset of mitosis, the two centrosomes ensure the assembly of a bipolar mitotic spindle and the strict segregation of sister chromatids between offspring cells, resulting in two cells with one centrosome and one genome set each. During nuclear formation in mammalian cell lines and X. laevis egg extracts, γstrings establish a nuclear protein boundary around chromatin that connects the cytoplasm and the nuclear compartment together throughout interphase (Figure 1 and Table 1) [6][17]. This chromatin-associated γstring boundary serves as a supporting scaffold for the formation of a NE around chromatin by facilitating the nucleation of lamin B1 during nuclear formation. Moreover, at the NE, γtubulin is associated with Ran and with nuclear pore proteins (Figure 1 and Table 1) [34][99]. In the nuclear compartment, mass spectrometry analyses of purified fractions of human nucleoli identified γtubulin at that location together with nucleolin, the most abundant RNA-binding protein at that site (Figure 1 and Table 1) [26][27][100,101]. At the G1/S transition, the phosphorylation of γtubulin on Ser131 and Ser385 regulates the recruitment of this protein to the nascent centriole, where it enables centrosome replication and also promotes the accumulation of γtubulin in the nucleus [41][12][37][36][42][43][44][45][85,88,100,106,108,109,110,111]. In human U2OS osteosarcoma cells, mutations in the GTP/magnesium-interacting residue Cys13 of γtubulin are cytotoxic [19][7][46][47][16,18,112,113], and treatment with either of the γtubulin GTPase binding domain inhibitors Citral Dimethyl Acetyl (CDA) and DiMethyl Fumarate (DMF,) disassembles γtubules [19][20][16,95]. These observations strongly suggest that the GTPase domain of γtubulin is essential for the dynamics of the γtubulin meshwork. The “where” and “when” characteristics of the self-polymerizing ability of γtubulin are most likely regulated by GTP acting together with phosphorylation-dependent changes in the conformation of γtubulin.
4. The Gamma-Tubulin Meshwork and Gene Transcription
In a genetic screen to identify proteins required for the proper functioning of homeotic genes in drosophila melanogaster, mutations in the brahma (brm) gene, a gene product related to the chromatin remodeling complex SWI/SNF, showed a genetic interaction with γTub23C (γtubulin1) mutations [48][114], suggesting a role of γtubulin1 in transcription. In this context, γtubulin interacts with the transcription factor family E2F in animals and plants (Figure 1 and Table 1) [12][28][49][88,102,103]. Indeed, nuclear γtubulin was found to bind to the DNA on the same DNA binding motif as E2F, leading to the view that the tumor suppressor retinoblastoma (RB)1 and γtubulin proteins complement each other in the regulation of gene expression [50][108]. The RB1/γtubulin signal network governs E2Fs, whose transcriptional activities induce the expression of the target genes that are indispensable for centrosome duplication and DNA replication [51][115]. Interestingly, besides interacting with E2Fs, RB1 can recruit remodeling factors, including histone deacetylases, members of the chromatin remodeling complex SWI/SNF, and DNA methyltransferase, aiding in the modification of the structure and organization of chromatin [52][53][54][116,117,118]. Altogether, these data indicate that γtubulin may be involved in the recruitment of DNA-remodeling factors.
In the nucleoli, electron microscopy experiments, performed by Hořejší and colleagues [37][100], showed that nucleolar γtubulin is present in the GC, where RNA transcription takes place (Table 1). It was also shown that, at that position, γtubulin is localized with the tumor suppressor C53, and this interaction is necessary for modulating the activity of C53 after treatment with DNA-damaging compounds, suggesting that the activity of γtubulin is necessary for DNA repair. In line with this assumption, γtubulin associates with Rad51, BReast CAncer type 1 susceptibility protein (BRCA)1, p53, Checkpoint Kinase (Chk)2, and Ataxia Telangiectasia and Ataxia Telangiectasia and Rad3-related protein (ATR), which are proteins that are involved in checkpoint activation and DNA repair [37][36][55][56][57][58][59][60][61][100,106,119,120,121,122,123,124,125].
5. The Gamma-Tubulin Meshwork and Nuclear Architecture
Depletion of γtubulin in Xenopus laevis egg extracts was shown to impair nuclear membrane formation [6][17]. Furthermore, live imaging of cells expressing γtubulin1 mutants, showed that impairment of the γstring boundary around chromatin led to the formation of chromatin empty nuclear-like structures, which collapsed into cytosolic lamin aggregates [6][17]. Moreover, in A. thaliana, γtubulin was found to be colocalized with SUN1 in the INM (Figure 1 and Table 1) [32][33]. It is worth noting that at the INM, both SUN1 and γtubulin interact with Samp1 and the nuclear lamina (Figure 1) [30][31][104,105]. Depletion of Samp1 impairs the proper recruitment of γtubulin to the mitotic spindle, revealing an important link between the interaction of nuclear proteins with γtubulin for the completion of cell division (Table 1) [30][104]. In the NE, γtubulin interacts with Mel-28/ELYS (Figure 1 and Table 1) [34][99], a Nup protein whose depletion impairs the proper formation of the NPC due to the inefficient recruitment of the Nup107-160 complex [62][126].
The fact that the PCM and γstrings are important sites that affect the nucleation and dynamics of MTs, actin filaments, and intermediate filaments (Table 1) [6][23][24][17,97,98], suggests that γstrings may function as a nucleating platform. With this in mind, the large number of γstrings associated with cellular membranes may support those membranes and provide a nucleating platform for cytoskeleton elements such as actin, MTs, or lamins, allowing them to mold and transmit signals into different cellular compartments (Figure 1 and Table 1). Accordingly, the interaction of γTURC with the Golgi membrane-linked GMAP-210 protein regulates the proper positioning and biogenesis of the Golgi apparatus [15][91]. In addition, γtubulin is associated with endosomes [13][89], and it is an important mitochondrial infrastructure that connects the mtDNA with the nuclear chromatin (Figure 1) [7][14][18,90]. Similarly, DNA-bound γstrings fasten the chromatin to the cytosol, and the site for the formation of a NE around the chromatin is marked by the transition between cytosolic and nuclear-associated γstrings [33][34][63][64][17,99,127,128]. Accordingly, in U2OS cells, treatment with either CDA or DMF (to increase the endogenous levels of the metabolite fumarate [49][65][103,129]), disassembled γstrings, disrupting the association between mitochondria and the nuclear compartment [7][18]. Altogether, these data support the notion that γtubulin is indispensable for structuring cellular compartments and coordinating the cytoplasm with the nuclear compartment.
Finally, in the cytoplasm, various γtubules emanating from a centrosome can intertwine, forming macro-γtubules, and the formation of those structures may influence the shape of the NE [5][14]. Altogether, findings may suggest that cytosolic γtubules, NE-inserted γstrings, and centrosomes work as structural docking sites for nuclear γtubulin, resulting in the recruitment of chromatin-remodeling factors necessary for the remodeling of the genome (Figure 1 and Figure 2).
6. The Gamma-Tubulin Meshwork and Cell Differentiation
Among the TUBG genes, TUBG1 is the most ubiquitous and predominantly expressed gene among all species, whereas TUBG2 and TUBG3 are expressed in the human brain and flowering plants, respectively. This implies that the expression of TUBG2 and TUBG3 isoforms are restricted to differentiated cells [4][66][84,87]. Indeed, in mice, TUBG1 is expressed in the cortex during embryonic development, but the expression of TUBG2 increases as the brain develops [67][130]. Differentiated neuroblastoma cells also express higher levels of γtubulin2, which further supports the idea that γtubulin1 and γtubulin2 have different functions during differentiation [68][90].
TUBG1 knock-out in mice is lethal. In contrast, TUBG2 knock-out mice are viable but exhibit some defects, including abnormalities in their circadian rhythm and painful stimulation [66][87]. In humans, mutations in the TUBG1 gene have been reported in children suffering from malformations related to cortical development [69][70][131,132]. Mice expressing the TUBG1 pathogenic variants suffer from cortex malformation and behavioral defects [67][130], proving the importance of γtubulin proteins in the development of the central nervous system.
A study conducted in isolated hippocampal neurons has shown that during the process of maturation, the centrosomal expression of γtubulin decreased after two weeks in culture [71][133]. Furthermore, in neurons, the nucleation of MTs does not rely on the centrosomal functions of γtubulin [72][73][134,135]. In general, in differentiated cells, there is a loss of MTOC activity in the centrosome and in the novel acquisition of MTOC activity at other cellular sites. This is partially achieved by altering the centrosomal localization of various proteins present in the PCM, which can be achieved in various ways [74][75][76][136,137,138]. For example, during differentiation of the mammalian epidermis, there is a transcriptional downregulation of genes encoding centrosomal proteins [77][139], whereas in neurons, the alternative splicing of the centrosome-targeting domain of the centrosomal protein ninein results in ninein dispersal [78][140]. In Drosophila oocytes, the MTOC pushes the nucleus, causing the formation of a groove on the NE, which results in the migration of the nucleus and the establishment of a dorsal-ventral axis [79][141].
7. Gamma-Tubulin in Cancer
As described above, γtubulin is involved in many crucial cellular processes, including cell proliferation and differentiation, processes that are hijacked during oncogenesis [80][142]. At the genomic level, few mutations or amplifications of TUBG genes have been reported in patients with cancer. According to the cbioportal [81][143], TUBG1 and TUBG2 have been observed to be amplified in rare subtypes of breast cancer (2 cases of adenoid cystic adenocarcinoma, N = 16 patients) and prostate cancer (Neuroendocrine Prostate Cancer, 19 cases, N = 114). Moreover, high levels of TUBG1 mRNA coincide with high levels of cell cycle-related genes in various tumor types, supporting the notion that γtubulin1 expression is necessary for proliferation [12][88]. Accordingly, in various tumors (retinoblastoma, bladder, breast, colorectal and small cell lung carcinoma (SCLC) tumors), γtubulin and RB1 moderate each other’s expression, and in the absence of γtubulin and RB1, the uncontrolled transcriptional activity of E2Fs upregulates apoptotic genes, causing cell death [12][50][88,108]. The RB pathway is one of the most well-described tumor suppressor pathways, and it is found to be highly mutated in a large spectrum of cancers [82][83][144,145]. Using the cbioportal [81][143], we found that among 31 subtypes of cancer, 30% exhibit defects in the RB pathway, with 7% of the patients exhibiting a loss of RB1 [82][144]. In brain tumors, high expression of γtubulin is associated with high-grade astrocytomas and glioblastomas, as compared to low-grade astrocytomas, which exhibited weaker γtubulin staining [84][146]. We can also point out that in medulloblastoma samples in areas of the tumors with high staining of the neuronal differentiation marker β3tubulin, the staining of γtubulin is low, whereas in tumor areas with low staining of β3tubulin, there is a strong co-staining of γtubulin and the proliferative marker proliferating cell nuclear antigen (PCNA) further confirming that there is a connection between γtubulin expression and highly proliferative tumors [85][147].
It is tempting to speculate that the high expression of γtubulin1 in proliferating cells may provide chromatin with more anchoring sites at the NE, which might assist in the maintenance of a plastic cell nucleus in proliferating cells, whereas, low expression of γtubulin1 will result in a stiffer nucleus that favors lineage-specific chromosome arrangements in less aggressive tumor cells [86][55].
Due to the functions of tubulins in the biology of the cell and, specifically, due to their involvement in mitotic spindle formation, since the 1960s, MTs have been the target of various chemotherapies that impair MT function, cell division, and angiogenesis, and thus reduce tumor growth [87][88][89][148,149,150]. These compounds are widely prescribed as antineoplastic drugs for a broad range of malignancies including lung, breast, gastric, esophageal, bladder, and prostate cancers, Kaposi’s sarcoma, and squamous cell carcinoma of the head and neck [87][149]. However, the effectiveness of MT-targeting drugs for cancer therapy is limited by drug resistance and severe side effects in treated patients [87][149]. The target for MT-targeting drugs is α/β tubulins heterodimers, but more recently, MT-targeting drugs that inhibit γtubulin have also been developed [89][151].
The recently discovered functions of γtubulin in the nucleus and their inverse correlation with the tumor suppressor RB1, together with the high expression of γtubulin in proliferating cells suggest that drugs specifically designed to inhibit the nuclear activity of γtubulin may pave the way for chemotherapies that target a broad range of malignant tumors but have no impact on healthy cells. The natural product, citral, affects αβ- and γtubulin activities [90][152], but the citral analog, CDA, specifically inhibits the nuclear activities of γtubulin without affecting MT dynamics [49][103]. CDA has been proven to have an in vivo antitumorigenic activity. Furthermore, the drug dimethyl fumarate (DMF) also targets the nuclear activities of γtubulin. DMF is a Food and Drug Administration (FDA)-approved drug for the treatment of multiple sclerosis and psoriasis [91][92][153,154] and, in addition, has been reported to diminish melanoma growth and metastasis in animal models [93][155]. Thus, based on the functions of γtubulin in the nucleus, the development of drugs that inhibit its nuclear activity may act specifically on tumor cells while sparing healthy tissue.
