Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Luca Ferrari.
Nipah virus (NiV) infection is a viral disease caused by a Henipavirus, belonging to the Paramyxoviridae family, responsible for a zoonosis. The course of the disease can be very serious and lead to death. NiV natural hosts are fruit bats (also known as megabats) belonging to the Pteropodidae family, especially those of the
Pteropus
genus. Natural infection in domestic animals has been described in farming pigs, horses, domestic and feral dogs and cats. Natural NiV transmission is possible intra-species (pig-to-pig, human-to-human) and inter-species (flying bat-to-human, pig-to-human, horse-to-human).
- Nipah virus
- pathology
- vaccines
- epidemiology
- diagnostics
- legislation
- pathogenesis
- immune response
- geographic information system
- spillover events
1. Introduction
Nipah virus (NiV) infection is a viral disease caused by a virus belonging to the Henipavirus genus, in the Paramyxoviridae family, responsible for a zoonosis whose main clinical manifestations in humans are respiratory and neurological. It can be defined as an emerging zoonosis. Zoonotic pathogens account for 60% of emerging infectious diseases, of which about 70% originate from wildlife [1]. The novel RNA paramyxovirus (genus Henipavirus), closely related to the Hendra virus, was named after the outbreak in the village of Sungai Nipah in the State of Negeri Sembilan (close to the Federal Territory of Kuala Lumpur), Malaysia, in which the virus was first isolated from a human patient in 1998 [2]. The course of the disease can be very serious and lead to death. The disease is also widely described in pigs, with clinical signs involving the respiratory and nervous systems [3]. The natural reservoir of the virus is represented by frugivorous bats called “flying foxes” belonging to the Pteropus genus. The main outbreaks of NiV infection have so far occurred in the geographical area of the carrier bats, i.e., in Malaysia, Singapore, Bangladesh and India, but cases were also described in the Philippines. The emergence of the virus and the zoonotic potential of transmission to animals and humans seem to be related to losses in the bats’ habitat [4]. The modes of transmission are different: Pteropus-swine-man, human contagion through the consumption of contaminated food, and inter-human contagion; direct bat-human contagion is also hypothesized. When outbreaks occur in pigs, the only possible measures are isolation, blocking of movements and killing of infected animals [5][6]. A definitive diagnosis can be performed by using molecular techniques, but serological tests are also available. NiV infection is an emerging and potentially dangerous disease whose spread must be curbed; currently, the main strategy is prevention via limiting contacts with reservoir species, using proper pig farming practices and improving the hygiene and feeding habits of some populations living in areas where the disease occurs [5]. Nowadays, drugs and vaccines and/or vaccine candidates in various species are scarcely employed or under investigation, therefore further studies are necessary to improve the knowledge about the interaction between NiV and the immune response, apply prevention methods, and develop effective prophylactic and curative therapies. In particular, due to the high risk in manipulating the virus itself in challenge animal studies, several experimental vaccines based on the main NiV immunogenic proteins or peptides have been developed mainly to investigate their immunogenicity, that is their ability to elicit a neutralizing antibody response potentially able to rapidly counteract the infection and induce a cell-mediated response to mediate viral clearance [7][8][9]. However, some experimental vaccines also elicited a response to varying degrees of clinical protection in model animals [3][10][11][12][13].
Furthermore, a better understanding of the spatial habitat preferences and diffusion of reservoir bats would be a useful contribution to their conservation and would help better prevent potential disease transmission [4]. Remote sensing (RS) and geographic information system (GIS) technologies have proven to be useful tools in investigating animal distribution in relation to environment/human settlements and diseases in wildlife [14][15], including the characteristics of the flying fox colonies potentially carrying NiV and their neighborhoods [4][16].
However, climate change, bringing an extreme increase in air temperature in tropical monsoon areas, has an adverse effect on the adaptive capacity of flying foxes to new environmental conditions, especially in relation to demography and species survival and thus NiV diffusion [17][18][19].
In Europe, the relative legislation refers to the generic management of communicable diseases, and specifically to the articles listed in the Regulation 2016/429 [20] about emerging diseases. In Malaysia, following the NiV outbreaks in pigs, a series of provisions aimed at containing the spread of the infection were planned, such as the strict isolation of the potential disease transmitters, the prohibition of the pig and meat trade, the establishment of an infected area and a protection area around the outbreaks, and the killing of infected pigs [2][21]. In India, following the recent cases in humans, the Indian National Centre for Disease Control (NCDC) established guidelines for the definition of suspected, probable and confirmed cases of NiV infection in order to detect and control the outbreaks early and subsequently regulate several public health activities aimed at containing the spread of the infection [3][22].
Therefore, a “One Health” approach, considering humans, domestic and peridomestic animals as well as the environment, is required to effectively control the disease.
2. Biological Features of the Nipah Virus (NiV)
NiV is a virus belonging to the Henipavirus genus, in the Paramyxoviridae family and Orthoparamyxovirinae subfamily [23][24]. The only other member of the genus endowed with pathogenicity and zoonotic potential is the Hendravirus (HeV) [25][26][27] identified in the 1990s following the death of several people after contact with infected equines in Australia [28]. At present, reports of spillover phenomena of the Henipavirus disease in humans are limited to Southeast Asia, although the widening of surveillance has allowed to identify these viruses in chiroptera sera in sub-Saharan Africa and Brazil [29][30][31][32][33][34]. Furthermore, in a recent study on sera from humans and bats in Cameroon, about 3–4% of human samples were found to be seropositive. Particularly, they were individuals who were involved in the butchering of bat meat [35]. Antibodies produced against NiV also cross-react with the HeV but not with other members of this family [36]. One of the most important biological characteristics of henipaviruses is the broad host spectrum both in vivo and in vitro [37]. Bats can be the main reservoir for some henipaviruses, which can cause a spontaneous disease in humans, pigs, horses, dogs and cats. Paramyxoviruses are enveloped pleomorphic viruses with an unsegmented single-stranded negative-sense RNA genome, encoding six structural and three accessory proteins [24][38][39]. These include the genome codes for nucleoprotein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), adhesion glycoprotein (G) and for large protein (L) or RNA polymerase in the order 3′-N-P-M-F-G-L-5′ [40][41][42]. The genome of some viruses of the Paramyxoviridae family codes for cell attachment proteins which can exert hemagglutination (H) and neuraminidase (N) functions. Specifically, the NiV G protein is involved in the adsorption phase but does not display H functions. In addition, the P gene also provides the three non-structural proteins V, W and C by means of mRNA editing mechanisms and alternative open reading frames (ORF) (Figure 1) [5][43].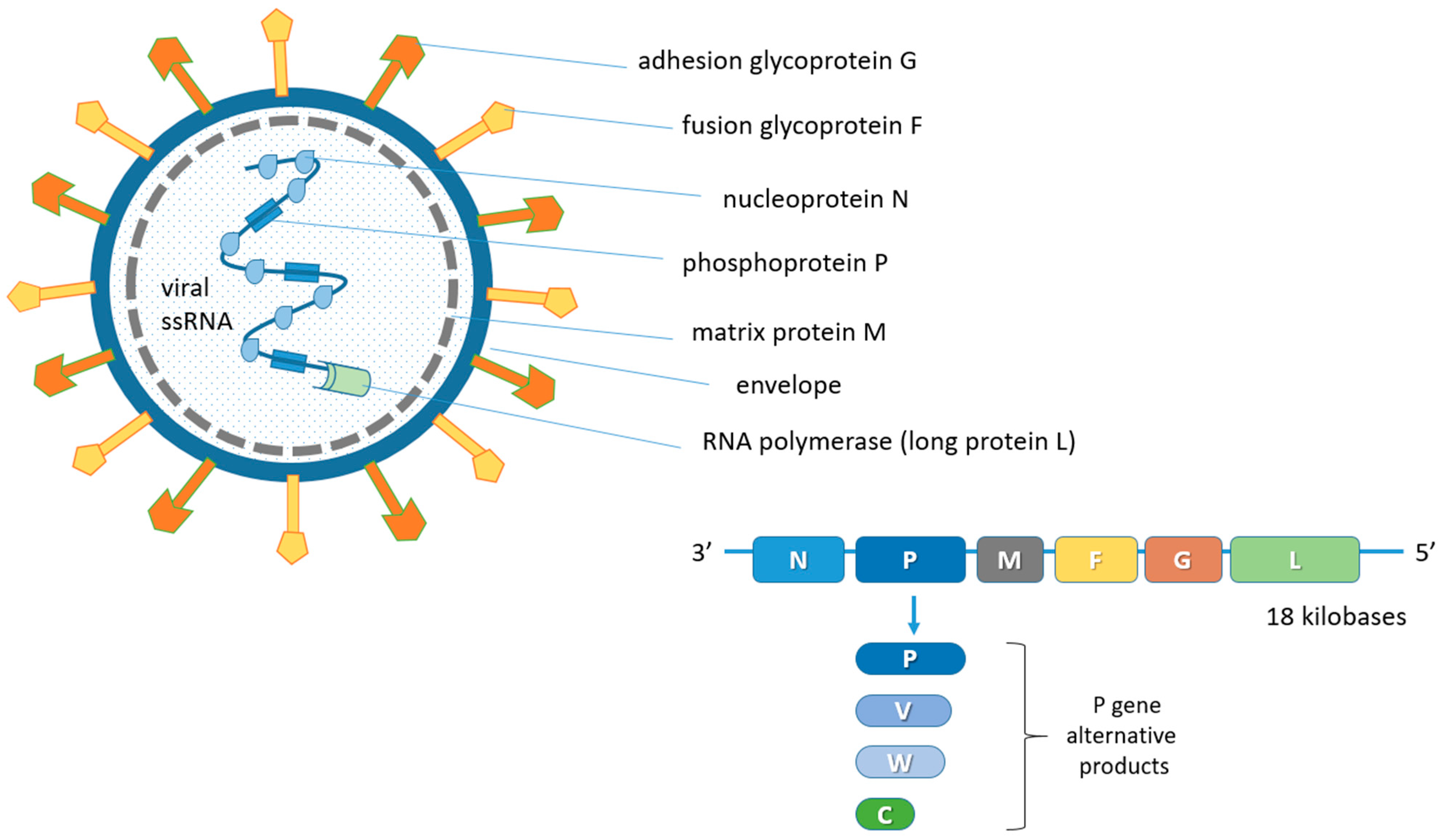
Figure 1. Schematic representation of the NiV structure and viral genome organization. N, P and L proteins interact with the unsegmented single-stranded negative-sense viral RNA genome forming the ribonucleoprotein (RNP) complex. The matrix M protein, used for viral assembly and budding, is associated with the inner side of the envelope which contains the G and F glycoproteins for adhesion to ephrin-B2 and -B3 receptors and fusion, respectively. The P gene products (V, W and C proteins) derive from mRNA editing and alternative open reading frames (ORF). The six coded genes are flanked by a 3′ leader and a 5′ trailer region.
3. Host Range
Natural hosts of NiV are fruit bats (also known as megabats) belonging to the Pteropodidae family, especially those of the Pteropus genus. Flying foxes are the fruit bats mostly involved in NiV spread, with direct shedding to receptive animals, human beings included, or through NiV-contaminated palm fruits (or palm sap) or biological bat matrixes (e.g., urine, faeces). Megabats represent NiV reservoirs in endemic geographical areas of Southeast Asia and sub-Saharan Africa.Natural infection in domestic animals has been described in farming pigs, horses, and domestic and feral cats. Natural NiV transmission can be intra-specific (pig-to-pig, human-to-human) and inter-specific (flying bat-to-human; pig-to-human and horse-to-human). Ruminants are spillover hosts in which NiV infection in the ovi-caprine is ascertained, while bovine is a species considered as NiV-permissive. Dogs are also susceptible to NiV infection, but dogs and cats do not seem to play a zoonotic role [65]. A study on NiV infection in peridomestic and feral cats in the Tioman island (Malaysia) pointed out that natural NiV infection is rare in cats and the zoonotic risk is classified as low [66].
4. Climate and Anthropogenic Influence on Pteropus Bats and NiV Spillover Events
Habitat loss is the greatest threat to wildlife and biodiversity. The loss and fragmentation of wildlife habitats can lead to increased contact among wildlife, domestic animals and humans, potentially leading to the emergence and spread of zoonotic diseases. Useful tools to understand spatial patterns and relationships among different variables, even apparently unrelated, are geographic information system (GIS) and remote sensing (RS) technologies. These geospatial technologies have been applied to gather information describing phenomena also in the veterinary field, such as the distribution of pathogen outbreaks and diseases in wildlife related to human behaviors and settlements as well as environmental factors and changes in climate in order to monitor and predict future pathogen spread and potential zoonotic outbreaks. These technologies help different categories of professionals and institutions to implement better management policies in a “One Health” perspective [4][14][15][16][72][73]. A better understanding of the Pteropus flying fox habitat preferences and diffusion would be a useful contribution to its conservation and would help better prevent potential disease transmission. In this regard, a study focused on the spatial characterization of colonies of flying foxes in Thailand using field observations, RS and GIS, showed that flying fox colonies are found particularly in locations surrounded by water, vegetation and controlled/protected religious areas. This aimed at mapping potential contact zones for bats, pigs and humans. High-risk areas for NiV zoonosis in pigs were proven to include the agricultural area around Bangkok where pig farms are numerous [4]. A recent GIS-based study on NiV-carrying bats in India and Bangladesh investigated the likely change in the distribution of Pteropus medius (based on data from 2015 onwards) under different future socioeconomic and environmental scenarios and the concurrent risk increase in NiV spillover events. It was predicted that the risk of NiV spillover events in India and Bangladesh will likely increase due to population growth and persistent environmental degradation. The adoption of rigorous public health measures in the areas predicted to be at major risk of NiV spillover from Pteropus medius to humans could reduce potential future spillovers and control future outbreaks [16]. Climate and anthropogenic changes can have important roles in the spread of viral infections to animals and humans [74]. Regarding NiV infection, it is thought that the emergence of the virus and the zoonotic transmission to susceptible animals and humans is related to losses in the reservoir bat’s habit. Fruit bats Pteropus populate the climate area of tropical monsoons, category Am, of the Köppen climate classification [75]. The tropical monsoon has a characteristic yearly constant temperature of 18 °C, as a minimum climate degree, and high rainfall. Tropical monsoon climate areas favor the maintenance of the tropical forest including fruit bats’ feed reserve and bats play a role (i.e., pollinators and seed dispersers) in maintaining vital tropical forest eco-systems [19][76]. Bats manage a wide range of body thermal regulation as homeotherms or poikilotherms. Flying fox bats are able to control body temperatures adapting to environment temperatures above 30 °C by decreasing the basal metabolic rate and entering a state of “torpor”. The bat, a nocturnal mammal, enters a state of controlled torpor during the hottest hours of the day, reducing basal metabolic activities to a minimum to overcome heat stress. The extreme environmental thermal events were studied in Australia and a significant rate of mortality was recorded in flying fox bats when the air temperature was ≥42.0 °C [17][18][19]. However, flying fox bat geographical distribution is highly dependent on food sources, a combination of nectar, pollen and fruit, especially those produced by Eucalyptus trees in woodlands and open forests [77][78][79]; therefore, a lack of food resources due to deforestation and climate change may disperse flying foxes outside their typical habitats with consequent increased levels of stress. Studies using urine cortisol concentrations to measure the physiological stress in Pteropid bats have pointed out that lower winter temperatures increase cortisol concentrations, which are associated with HeV excretion in flying foxes [80][81]. A similar condition may occur with NiV. Another aspect is the body condition score (BCS) that is related to the nutritional status. Poor body condition is associated with increased seroconversion and HeV infection risk in flying foxes. This increases the urinary excretion of viable virus, with an increased risk of transmission to horses, the main host of the HeV disease [82][83]. Specific weather patterns have been associated with spillover events. A number of studies have consistently demonstrated associations between dry conditions and spillover events of HeV [1][79][84][85][86][87]. El Niño cycles (warmer and drier conditions) in spring/summer correlates with spillover events in autumn/winter [79][84]. El Niño and drought events are predicted to occur more frequently in the future due to climate change [88][89]. Therefore, these extreme weather conditions coupled with the cascading effects on flying foxes may increase the risk and frequency of HeV spillover events/outbreaks and expansion into new areas. Field studies carried out in Malaysia have suggested a complex interaction of several factors that might have triggered the 1997–1998 outbreak: the reduction of the inhabitants of the reservoirs that are linked to deforestation for commercial and agricultural purposes, prolonged drought driven by the strong El Niño Southern Oscillation (ENSO) phenomenon, forest fires caused by man, intense promiscuity of crops and pig farms and overcrowding of pigsties. All this led to a greater probability of virus spillover from its natural reservoir to the pig and consequently to humans [90]. Regarding human behaviors and NiV infection, in a qualitative study in two districts of Bangladesh, interviews were conducted with 30 bat hunters who hunted bats primarily for consumption, to understand the process and reasons for hunting bats and their perception about bats and bat-borne disease. It was reported that bat meat is prepared for personal consumption and to be sold in neighboring communities. Live bats are also sold to traditional healers. Many informants (53%) reported being bitten by bats. Some informants reported hearing about a disease from bat-contaminated date palm sap, and NiV was mentioned, but they did not believe that bats could spread a disease to humans because of the absence of disease in their community that have been hunting and consuming bats for generations. However, the close bat–human interaction reported in this study highlights a risk of pathogen spillover [91]. Also, the trade of flying bat meat in markets in Southeast Asia may represent a source of potential infection. For instance, north and southeast areas of the Sulawesi Island in Indonesia are dynamic sites for the wildlife trade of hunted species in wild meat markets, including large amounts of flying foxes that roost are under hunting pressure. However, the current rates of flying fox harvests are significantly unstainable and the impact of this trade on local ecosystems remains mostly unknown [92].References
- Yuen, K.Y.; Fraser, N.S.; Henning, J.; Halpin, K.; Gibson, J.S.; Betzien, L.; Stewart, A.J. Hendra virus: Epidemiology dynamics in relation to climate change, diagnostic tests and control measures. One Health 2021, 12, 100207.
- Mohd Nor, M.N.; Gan, C.H.; Ong, B.L. Nipah virus infection of pigs in peninsular Malaysia. Rev. Sci. Tech. 2000, 19, 160–165.
- Aditi; Shariff, M. Nipah virus infection: A review. Epidemiol. Infect. 2019, 147, e95.
- Thanapongtharm, W.; Linard, C.; Wiriyarat, W.; Chinsorn, P.; Kanchanasaka, B.; Xiao, X.; Biradar, C.; Wallace, R.G.; Gilbert, M. Spatial characterization of colonies of the flying fox bat, a carrier of Nipah virus in Thailand. BMC Vet. Res. 2015, 11, 81.
- Singh, R.K.; Dhama, K.; Chakraborty, S.; Tiwari, R.; Natesan, S.; Khandia, R.; Munjal, A.; Vora, K.S.; Latheef, S.K.; Karthik, K.; et al. Nipah virus: Epidemiology, pathology, immunobiology and advances in diagnosis, vaccine designing and control strategies—A comprehensive review. Vet. Q. 2019, 39, 26–55.
- Pillai, V.S.; Krishna, G.; Veettil, M.V. Nipah virus: Past outbreaks and future containment. Viruses 2020, 12, 465.
- Satterfield, B.A.; Dawes, B.E.; Milligan, G.N. Status of vaccine research and development of vaccines for Nipah virus. Vaccine 2016, 34, 2971–2975.
- Devnath, P.; Wajed, S.; Chandra Das, R.; Kar, S.; Islam, I.; Masud, H.M.A.A. The pathogenesis of Nipah virus: A review. Microb. Pathog. 2022, 170, 105693.
- Pedrera, M.; Macchi, F.; McLean, R.K.; Franceschi, V.; Thakur, N.; Russo, L.; Medfai, L.; Todd, S.; Tchilian, E.Z.; Audonnet, J.C.; et al. Bovine herpesvirus-4-vectored delivery of Nipah virus glycoproteins enhances T cell immunogenicity in pigs. Vaccines 2020, 8, 115.
- Mungall, B.A.; Middleton, D.; Crameri, G.; Bingham, J.; Halpin, K.; Russell, G.; Green, D.; McEachern, J.; Pritchard, L.I.; Eaton, B.T.; et al. Feline model of acute Nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J. Virol. 2006, 80, 12293–12302.
- Pallister, J.A.; Klein, R.; Arkinstall, R.; Haining, J.; Long, F.; White, J.R.; Payne, J.; Feng, Y.R.; Wang, L.F.; Broder, C.C.; et al. Vaccination of ferrets with a recombinant G glycoprotein subunit vaccine provides protection against Nipah virus disease for over 12 months. Virol. J. 2013, 10, 237.
- Weingartl, H.M.; Berhane, Y.; Caswell, J.L.; Loosmore, S.; Audonnet, J.C.; Roth, J.A.; Czub, M. Recombinant Nipah virus vaccines protect pigs against challenge. J. Virol. 2006, 80, 7929–7938.
- Yoneda, M.; Georges-Courbot, M.C.; Ikeda, F.; Ishii, M.; Nagata, N.; Jacquot, F.; Raoul, H.; Sato, H.; Kai, C. Recombinant measles virus vaccine expressing the Nipah virus glycoprotein protects against lethal Nipah virus challenge. PLoS ONE 2013, 8, e58414.
- Orusa, T.; Orusa, R.; Viani, A.; Carella, E.; Borgogno Mondino, E. Geomatics and EO data to support wildlife diseases assessment at landscape level: A pilot experience to map infectious keratoconjunctivitis in Chamois and phenological trends in Aosta Valley (NW Italy). Remote Sens. 2020, 12, 3542.
- Carella, E.; Orusa, T.; Viani, A.; Meloni, D.; Borgogno Mondino, E.; Orusa, R. An integrated, tentative remote-sensing approach based on NDVI entropy to model canine distemper virus in wildlife and to prompt science-based management policies. Animals 2022, 12, 1049.
- Kumar, V.B.; Rooney, N.; Carr, A. Nipah virus from bats—Another potential pandemic? Risk mapping the impact of anthropogenic and climate change on the transmission of Nipah virus infection to humans. medRxiv 2022.
- Welbergen, J.A.; Klose, S.M.; Markus, N.; Eby, P. Climate change and the effects of temperature extremes on Australian flying-foxes. Proc. Biol. Sci. 2008, 275, 419–425.
- Ratnayake, H.U.; Kearney, M.R.; Govekar, P.; Karoly, D.; Welbergen, J.A. Forecasting wildlife die-offs from extreme heat events. Anim. Conserv. 2019, 22, 386–395.
- Diengdoh, V.L.; Ondei, S.; Hunt, M.; Brook, B.W. Predicted impacts of climate change and extreme temperature events on the future distribution of fruit bat species in Australia. Glob. Ecol. Conserv. 2022, 37, e02181.
- European Union (EU). Regulation (EU) 2016/429 of the European Parliament and of the Council of 9 March 2016 on Transmissible Animal Diseases and Amending and Repealing Certain Acts in the Area of Animal Health (‘Animal Health Law’). Off. J. 2016. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32016R0429&from=EN (accessed on 22 December 2022).
- Aziz, B.J.; Azri, B.A. Nipah virus infection—Malaysia experience. In Proceedings of the World Organization of Animal Health (OIE) Conference WILDLIFE ACTES 2011; Available online: https://www.woah.org/fileadmin/Home/eng/Conferences_Events/sites/WILDLIFE_ACTES_2011/Presentations/S1_3_AzriBinAdzhar.pdf (accessed on 22 December 2022).
- National Centre for Disease Control (NCDC) India. Available online: https://ncdc.gov.in/ (accessed on 22 December 2022).
- Chua, K.B.; Bellini, W.J.; Rota, P.A.; Harcourt, B.H.; Tamin, A.; Lam, S.K.; Ksiazek, T.G.; Rollin, P.E.; Zaki, S.R.; Goldsmith, C.S. Nipah virus: A recently emergent deadly paramyxovirus. Science 2000, 288, 1432–1435.
- Li, T.; Shen, Q.T. Insights into paramyxovirus nucleocapsids from diverse assemblies. Viruses 2021, 13, 2479.
- Middleton, D. Hendra virus. Vet. Clin. N. Am. Equine Pract. 2014, 30, 579–589.
- Middleton, D.; Pallister, J.; Klein, R.; Feng, Y.R.; Haining, J.; Arkinstall, R.; Frazer, L.; Huang, J.A.; Edwards, N.; Wareing, M.; et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis. 2014, 20, 372–379.
- Enchèry, F.; Horvat, B. Understanding the interaction between Henipaviruses and their natural host, fruit bats: Paving the way toward control of highly lethal infection in humans. Int. Rev. Immunol. 2017, 36, 108–121.
- Eaton, B.T.; Broder, C.C.; Middleton, D.; Wang, L.F. Hendra and Nipah viruses: Different and dangerous. Nat. Rev. Microbiol. 2006, 4, 23–35.
- Iehlé, C.; Razafitrimo, G.; Razainirina, J.; Andriaholinirina, N.; Goodman, S.M.; Faure, C.; Georges-Courbot, M.C.; Rousset, D.; Reynes, J.M. Henipavirus and Tioman virus antibodies in pteropodid bats, Madagascar. Emerg. Infect. Dis. 2007, 13, 159–161.
- Hayman, D.T.S.; Suu-Ire, R.; Breed, A.C.; McEachern, J.A.; Wang, L.; Wood, J.L.N.; Cunningham, A.A. Evidence of Henipavirus infection in West African fruit bats. PLoS ONE 2008, 3, e2739.
- Drexler, J.F.; Corman, V.M.; Gloza-Rausch, F.; Seebens, A.; Annan, A.; Ipsen, A.; Kruppa, T.; Müller, M.A.; Kalko, E.K.V.; Adu-Sarkodie, Y.; et al. Henipavirus RNA in African bats. PLoS ONE 2009, 4, e6367.
- Hayman, D.T.S.; Wang, L.F.; Barr, J.; Baker, K.S.; Suu-Ire, R.; Broder, C.C.; Cunningham, A.A.; Wood, J.L.N. Antibodies to Henipavirus or henipa-like viruses in domestic pigs in Ghana, West Africa. PLoS ONE 2011, 6, e25256.
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Cottontail, V.M.; Rasche, A.; Yordanov, S.; et al. Bats host major mammalian paramyxoviruses. Nat. Commun. 2012, 3, 796.
- Mbu’u, C.M.; Mbacham, W.F.; Gontao, P.; Sado Kamdem, S.L.; Nlôga, A.M.N.; Groschup, M.H.; Wade, A.; Fischer, K.; Balkema-Buschmann, A. Henipaviruses at the interface between bats, livestock and human population in Africa. Vector-Borne Zoonotic Dis. 2019, 19, 455–465.
- Pernet, O.; Schneider, B.S.; Beaty, S.M.; LeBreton, M.; Yun, T.E.; Park, A.; Zachariah, T.T.; Bowden, T.A.; Hitchens, P.; Ramirez, C.M.; et al. Evidence for Henipavirus spillover into human populations in Africa. Nat. Commun. 2014, 5, 5342.
- Zhu, Z.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Bishop, K.A.; Choudhry, V.; Mungall, B.A.; Feng, Y.R.; Choudhary, A.; Zhang, M.Y.; et al. Potent neutralization of Hendra and Nipah viruses by human monoclonal antibodies. J. Virol. 2006, 80, 891–899.
- Bellini, W.J.; Harcourt, B.H.; Bowden, N.; Rota, P.A. Nipah virus: An emergent paramyxovirus causing severe encephalitis in humans. J. Neurovirol. 2005, 11, 481–487.
- Lamb, R.A.; Parks, G.D. Paramyxoviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2006; pp. 1449–1496.
- El Najjar, F.; Schmitt, A.P.; Dutch, R.E. Paramyxovirus glycoprotein incorporation, assembly and budding: A three way dance for infectious particle production. Viruses 2014, 6, 3019–3054.
- Maisner, A.; Neufeld, J.; Weingartl, H. Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb. Haemost. 2009, 102, 1014–1023.
- Aguilar, H.C.; Henderson, B.A.; Zamora, J.L.; Johnston, G.P. Paramyxovirus glycoproteins and the membrane fusion process. Curr. Clin. Microbiol. Rep. 2016, 3, 142–154.
- Walpita, P.; Cong, Y.; Jahrling, P.B.; Rojas, O.; Postnikova, E.; Yu, S.; Johns, L.; Holbrook, M.R. A VLP-based vaccine provides complete protection against Nipah virus challenge following multiple-dose or single-dose vaccination schedules in a hamster model. NPJ Vaccines 2017, 2, 21.
- Wang, L.; Harcourt, B.H.; Yu, M.; Tamin, A.; Rota, P.A.; Bellini, W.J.; Eaton, B.T. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 2001, 3, 279–287.
- Harcourt, B.H.; Tamin, A.; Ksiazek, T.G.; Rollin, P.E.; Anderson, L.J.; Bellini, W.J.; Rota, P.A. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 2000, 271, 334–349.
- Harcourt, B.H.; Lowe, L.; Tamin, A.; Liu, X.; Bankamp, B.; Bowden, N.; Rollin, P.E.; Comer, J.A.; Ksiazek, T.G.; Hossain, M.J.; et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg. Infect. Dis. 2005, 11, 1594–1597.
- de Wit, E.; Munster, V.J. Nipah virus emergence, transmission, and pathogenesis. In Global Virology I—Identifying and Investigating Viral Diseases; Shapshak, P., Sinnott, J., Somboonwit, C., Kuhn, J., Eds.; Springer: New York, NY, USA, 2015; pp. 125–146.
- de Wit, E.; Munster, V.J. Animal models of disease shed light on Nipah virus pathogenesis and transmission. J. Pathol. 2015, 235, 196–205.
- Angeletti, S.; Presti, A.L.; Cella, E.; Ciccozzi, M. Molecular epidemiology and phylogeny of Nipah virus infection: A mini review. Asian Pac. J. Trop. Med. 2016, 9, 630–634.
- AbuBakar, S.; Chang, L.Y.; Ali, A.R.; Sharifah, S.H.; Yusoff, K.; Zamrod, Z. Isolation and molecular identification of Nipah virus from pigs. Emerg. Infect. Dis. 2004, 10, 2228–2230.
- Liew, Y.J.M.; Ibrahim, P.A.S.; Ong, H.M.; Chong, C.N.; Tan, C.T.; Schee, J.P.; Gómez Román, R.; Cherian, N.G.; Wong, W.F.; Chang, L.Y. The immunobiology of Nipah virus. Microorganisms 2022, 10, 1162.
- Tan, C.T.; Chua, K.B. Nipah virus encephalitis. Curr. Infect. Dis. Rep. 2008, 10, 315–320.
- Vidal, S.; Curran, J.; Kolakofsky, D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 1990, 64, 239–246.
- Steward, M.; Vipond, I.B.; Millar, N.S.; Emmerson, P.T. RNA editing in Newcastle disease virus. J. Gen. Virol. 1993, 74, 2539–2547.
- Delenda, C.; Taylor, G.; Hausmann, S.; Garcin, D.; Kolakofsky, D. Sendai viruses with altered P, V, and W protein expression. Virology 1998, 242, 327–337.
- Yu, M.; Hansson, E.; Langedijk, J.P.; Eaton, B.T.; Wang, L.F. The attachment protein of Hendra virus has high structural similarity but limited primary sequence homology compared with viruses in the genus Paramyxovirus. Virology 1998, 251, 227–233.
- Reynes, J.M.; Counor, D.; Ong, S.; Faure, C.; Seng, V.; Molia, S.; Walston, J.; Georges-Courbot, M.C.; Deubel, V.; Sarthou, J.L. Nipah virus in Lyle’s flying foxes, Cambodia. Emerg. Infect. Dis. 2005, 11, 1042–1047.
- Chadha, M.S.; Comer, J.A.; Lowe, L.; Rota, P.A.; Rollin, P.E.; Bellini, W.J.; Ksiazek, T.G.; Mishra, A. Nipah virus-associated encephalitis outbreak, Siliguri, India. Emerg. Infect. Dis. 2006, 12, 235–240.
- Bonaparte, M.I.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Mungall, B.A.; Bishop, K.A.; Choudhry, V.; Dimitrov, D.S.; Wang, L.F.; Eaton, B.T. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10652–10657.
- Negrete, O.A.; Levroney, E.L.; Aguilar, H.C.; Bertolotti-Ciarlet, A.; Nazarian, R.; Tajyar, S.; Lee, B. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 2005, 436, 401–405.
- Negrete, O.A.; Wolf, M.C.; Aguilar, H.C.; Enterlein, S.; Wang, W.; Mühlberger, E.; Su, S.V.; Bertolotti-Ciarlet, A.; Flick, R.; Lee, B. Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2006, 2, e7.
- Liebl, D.J.; Morris, C.J.; Henkemeyer, M.; Parada, L.F. mRNA expression of ephrins and Eph receptor tyrosine kinases in the neonatal and adult mouse central nervous system. J. Neurosci. Res. 2003, 71, 7–22.
- Zimmer, M.; Palmer, A.; Köhler, J.; Klein, R. Ephb-ephrinB bi-directional endocytosis terminates adhesion allowing contact-mediated repulsion. Nat. Cell Biol. 2003, 5, 869–878.
- Bossart, K.N.; Tachedjian, M.; McEachern, J.A.; Crameri, G.; Zhu, Z.; Dimitrov, D.S.; Broder, C.C.; Wang, L.F. Functional studies of host-specific ephrin-B ligands as Henipavirus receptors. Virology 2008, 372, 357–371.
- Hassan, M.Z.; Sazzad, H.M.S.; Luby, S.P.; Sturm-Ramirez, K.; Bhuiyan, M.U.; Rahman, M.Z.; Islam, M.M.; Ströher, U.; Sultana, S.; Kafi, M.A.H.; et al. Nipah virus contamination of hospital surfaces during outbreaks, Bangladesh, 2013–2014. Emerg. Infect. Dis. 2018, 24, 15–21.
- Nikolay, B.; Salje, H.; Hossain, M.J.; Khan, A.K.M.D.; Sazzad, H.M.S.; Rahman, M.; Daszak, P.; Ströher, U.; Pulliam, J.R.C.; Kilpatrick, A.M.; et al. Transmission of Nipah virus—14 Years of investigations in Bangladesh. N. Engl. J. Med. 2019, 380, 1804–1814.
- Epstein, J.H.; Rahman, S.A.; Zambriski, A.; Halpin, K.; Meehan, G.; Jamaluddin, A.A.; Hassan, S.S.; Field, H.E.; Hyatt, A.D.; Daszak, P. Henipavirus Ecology Research Group. Feral cats and risk for Nipah virus transmission, Emerg. Infect. Dis. 2006, 12, 1178–1179.
- World Organization for Animal Health (OIE). Nipah Virus Infection (Wild Animals). Available online: https://www.woah.org/en/disease/nipah-virus-infection-wild-animals/ (accessed on 22 December 2022).
- World Organization for Animal Health (OIE). Henipaviruses (Nipah viruses) (Infection with). Available online: https://www.woah.org/app/uploads/2022/02/henipaviruses-nipah-viruses-infection-with.pdf (accessed on 22 December 2022).
- Eshaghi, M.; Tan, W.S.; Mohodin, T.B.; Yusoff, K. Nipah virus glycoprotein: Production in baculovirus and application in diagnosis. Virus Res. 2004, 106, 71–76.
- Weingartl, H.M. Hendra and Nipah viruses: Pathogenesis, animal models and recent breakthroughs in vaccination. Vaccine 2015, 5, 59–74.
- Ching, P.K.G.; de los Reyes, V.C.; Sucaldito, M.N.; Tayag, E.; Columna-Vingno, A.B.; Malbas, F.F., Jr.; Bolo, G.C., Jr.; Sejvar, J.J.; Eagles, D.; Playford, G.; et al. Outbreak of Henipavirus infection, Philippines, 2014. Emerg. Infect. Dis. 2015, 21, 328–331.
- Anyamba, A.; Chretien, J.P.; Britch, S.C.; Soebiyanto, R.P.; Small, J.L.; Jepsen, R.; Forshey, B.M.; Sanchez, J.L.; Smith, R.D.; Harris, R.; et al. Global disease outbreaks associated with the 2015-2016 El Niño event. Sci. Rep. 2019, 9, 1930.
- Orusa, T.; Borgogno Mondino, E. Exploring short-term climate change effects on rangelands and broad-leaved forests by free satellite data in Aosta Valley (Northwest Italy). Climate 2021, 9, 47.
- Carlson, C.J.; Albery, G.F.; Merow, C.; Trisos, C.H.; Zipfel, C.M.; Eskew, E.A.; Olival, K.J.; Ross, N.; Bansal, S. Climate change increases cross-species viral transmission risk. Nature 2022, 607, 555–562.
- Translated by Volken, E.; Brönnimann, S. 1884. Köppen, W. “Die Wärmezonen der Erde, nach der Dauer der heissen, gemässigten und kalten Zeit und nach der Wirkung der Wärme auf die organische Welt betrachtet” . Meteorol. Z. 2011, 20, 351–360.
- Mickleburgh, S.P.; Hutson, A.M.; Racey, P.A. Old World Fruit Bats—An Action Plan for Their Conservation; International Union for Conservation of Nature and Natural Resources (IUCN): Gland, Switzerland, 1992; ISBN 2-8317-0055-8. Available online: https://portals.iucn.org/library/efiles/documents/1992-034.pdf (accessed on 22 December 2022).
- Palmer, C.; Price, O.F.; Bach, C. Foraging ecology of the black flying fox (Pteropus alecto) in the seasonal tropics of the Northern Territory, Australia. Wildl. Res. 2000, 27, 169–178.
- Courts, S. Dietary strategies of old world fruit bats (Megachiroptera, Pteropodidae): How do they obtain sufficient protein? Mammal Rev. 1998, 28, 185–193.
- Giles, J.R.; Eby, P.; Parry, H.; Peel, A.J.; Plowright, R.K.; Westcott, D.A.; McCallum, H. Environmental drivers of spatiotemporal foraging intensity in fruit bats and implications for Hendra virus ecology. Sci. Rep. 2018, 8, 9555.
- Edson, D.; Field, H.; McMichael, L.; Jordan, D.; Kung, N.; Mayer, D.; Smith, C. Flying-fox roost disturbance and Hendra virus spillover risk. PLoS ONE 2015, 10, e0125881.
- McMichael, L.; Edson, D.; Smith, C.; Mayer, D.; Smith, I.; Kopp, S.; Meers, J.; Field, H. Physiological stress and Hendra virus in flying-foxes (Pteropus spp.), Australia. PLoS ONE 2017, 12, e0182171.
- Edson, D.; Peel, A.J.; Huth, L.; Mayer, D.G.; Vidgen, M.E.; McMichael, L.; Broos, A.; Melville, D.; Kristoffersen, J.; de Jong, C.; et al. Time of year, age class and body condition predict Hendra virus infection in Australian black flying foxes (Pteropus alecto). Epidemiol. Infec. 2019, 147, e240.
- Plowright, R.K.; Field, H.E.; Smith, C.; Divljan, A.; Palmer, C.; Tabor, G.; Daszak, P.; Foley, J.E. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc. Biol. Sci. 2008, 275, 861–869.
- McFarlane, R.; Becker, N.; Field, H. Investigation of the climatic and environmental context of Hendra virus spillover events 1994–2010. PLoS ONE 2011, 6, e28374.
- Páez, D.J.; Giles, J.; McCallum, H.; Field, H.; Jordan, D.; Peel, A.J.; Plowright, R.K. Conditions affecting the timing and magnitude of Hendra virus shedding across pteropodid bat populations in Australia. Epidemiol. Infec. 2017, 145, 3143–3153.
- Martin, G.; Yanez-Arenas, C.; Chen, C.; Plowright, R.K.; Webb, R.J.; Skerratt, L.F. Climate change could increase the geographic extent of Hendra virus spillover risk. EcoHealth 2018, 15, 509–525.
- Martin, G.; Yanez-Arenas, C.; Plowright, R.K.; Chen, C.; Roberts, B.; Skerratt, L.F. Hendra virus spillover is a bimodal system driven by climatic factors. EcoHealth 2018, 15, 526–542.
- Timmermann, A.; Oberhuber, J.; Bacher, A.; Esch, M.; Latif, M.; Roeckner, E. Increased El Niño frequency in a climate model forced by future greenhouse warming. Nature 1999, 398, 694–697.
- Mpelasoka, F.; Hennessy, K.; Jones, R.; Bates, B. Comparison of suitable drought indices for climate change impacts assessment over Australia towards resource management. Int. J. Climatol. 2008, 28, 1283–1292.
- Chua, K.B.; Koh, C.L.; Hooi, P.S.; Wee, K.F.; Khong, J.H.; Chua, B.H.; Chan, Y.P.; Lim, M.E.; Lam, S.K. Isolation of Nipah virus from Malaysian flying-foxes. Microbes Infect. 2002, 4, 145–151.
- Nahar, N.; Asaduzzaman, M.; Mandal, U.K.; Rimi, N.A.; Gurley, E.S.; Rahman, M.; Garcia, F.; Zimicki, S.; Sultana, R.; Luby, S.P. Hunting bats for human consumption in Bangladesh. EcoHealth 2020, 17, 139–151.
- Latinne, A.; Saputro, S.; Kalengkongan, J.; Kowel, C.L.; Gaghiwu, L.; Ransaleleh, T.A.; Nangoy, M.J.; Wahyuni, I.; Kusumaningrum, T.; Safari, D.; et al. Characterizing and quantifying the wildlife trade network in Sulawesi, Indonesia. Glob. Ecol. Conserv. 2020, 21, e00887.
- Latinne, A.; Saputro, S.; Kalengkongan, J.; Kowel, C.L.; Gaghiwu, L.; Ransaleleh, T.A.; Nangoy, M.J.; Wahyuni, I.; Kusumaningrum, T.; Safari, D.; et al. Characterizing and quantifying the wildlife trade network in Sulawesi, Indonesia. Glob. Ecol. Conserv. 2020, 21, e00887.
More
