1. The PPARG Gene
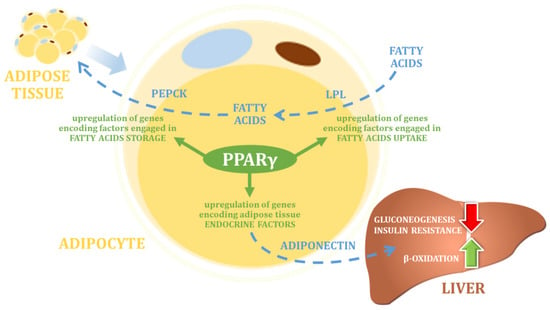
Within the PPAR protein family, peroxisome proliferator-activated receptor gamma (PPARγ) is one of the most frequently described factors in the context of its role in regulating metabolic changes that may affect body weight changes. PPARγ protein is expressed in adipocytes (where its expression is highest) in white and brown adipose tissue, and in the large intestine and spleen. PPARγ plays a key role in the regulation of adipocyte differentiation and adipogenesis, as well as in the regulation of lipid biosynthesis and lipoprotein metabolism, and thus in the control of energy balance (
Figure 12)
[1][47]. Biochemical studies revealed that PPARγ agonists promote a flow of fatty acids into adipose tissue and away from skeletal muscle and liver under physiological conditions
[2][3][4][5][48,49,50,51]. These processes lead to a decrease in fatty acid metabolism, and consequently to an increase in glucose utilization via the Randle cycle in muscles
[6][52]. Most of these effects are mediated in a complicated network via insulin-dependent signaling pathways
[7][53].
In humans, the PPARγ protein is encoded by the
PPARG gene, which is located on the chromosome 3 at position 3p25.2
[8][54]. Analyses of the genetic expression process have shown that in the case of the
PPARG gene, the phenomenon of the alternative reading of its sequence using different promoters (including those located within the gene) is observed, combined with the alternative splicing of primary transcripts (hnRNA), leading to the formation of different types of mature mRNA molecules (labeled with symbols from mRNAγ1 to mRNAγ7), and consequently, to the formation of many isoforms of the PPARγ protein (referred to as PPARγ1–PPARγ7)
[9][10][11][55,56,57]. Each of these isoforms is characterized by a well-defined expression pattern, differential tissue distribution, and specific activity. The presence of so many variants of the PPARγ protein means that this transcription factor exerts a very broad range of functions in the human body.
Numerous reports suggest that the functional differentiation of the PPARγ protein may be important for maintaining a healthy body weight
[12][13][14][15][16][17][58,59,60,61,62,63]. PPARγ is known to be a modulator of insulin-dependent signaling pathways—indicating that it may act through a variety of complex metabolic networks
[18][19][20][21][22][64,65,66,67,68].
AuthorsWe now know with certainty that PPARγ can sensitize skeletal muscle and liver to insulin
[23][24][25][69,70,71], but
thwe
y do not know the full mechanism that is responsible for this effect. One of the most surprising observations (from a study on rodents) is that Pparγ can sensitize tissues to insulin, both via the supraphysiological stimulation of Pparγ with antidiabetic drugs, and via a decrease in Pparγ activity, resulting from a complete knockout of the
Pparg gene
[26][72], or (from a study on humans) via changes in its functionality due to a specific polymorphic variant of the
PPARG gene
[27][73]. People who are carriers of certain polymorphic forms (alleles) of the
PPARG gene may be somewhat “metabolically burdened” because the PPARγ protein that they produce is less or more active as a stimulator of the expression of certain genes involved in fatty acid and glucose metabolism, or the regulation of insulin-dependent signaling pathways
[27][28][73,74]. Indeed,
PPARG may contribute to obesity
[14][60] by controlling food intake and appetite, not only through changes in insulin sensitivity
[26][72], but also by regulating the transcription of the leptin gene
[29][75], which is a key regulator of the central control of eating behavior. Other studies suggest that the presence of certain alleles in the
PPARG gene can lead to obesity
[14][60], insulin resistance, and type 2 diabetes
[30][76] or metabolic syndrome
[31][77].
Figure 12. The main physiological function of PPARγ (own elaboration, after: Skat-Rørdam et al. 2018
[32][78]). PEPCK—Phosphoenolpyruvate carboxykinase; LPL—lipoprotein lipase.
The most commonly studied polymorphic variant described as mediating nutritional effects on weight loss and maintenance is the C34G substitution (rs1801282), located in exon B, which is an integral part of the
PPARG gene
[33][79]. As a result of this nucleotide substitution at position 12 of the encoded PPARγ2 protein, alanine occurs instead of proline (Pro12Ala polymorphism). The presence of alanine at position 12 (12Ala allele) is associated with a reduced binding affinity of the PPARγ2 protein to the promoter regions of the PPARγ-controlled target genes, resulting in their much weaker activation
[34][35][80,81]. The functional significance of the amino acid modification Pro12Ala, in the PPARγ2 protein, results from its localization within the PPARγ2 molecule, namely within the AF-1 domain of the amino terminus of the PPARγ2 protein, which controls ligand-independent transcriptional activity
[36][82]. It is suggested that the Pro12Ala change in the AF-1 domain may indirectly facilitate phosphorylation and/or sumoylation processes, which reduce the potential of liganded PPARγ2 to activate the transcription of its target genes
[7][37][38][39][53,83,84,85].
As mentioned above, PPARγ regulates adipocyte differentiation and controls body fat storage. Therefore, it is natural that the differentiation of the
PPARG gene, and in particular, the functional significance of the Pro12Ala variants in relation to body weight measured by BMI and susceptibility to obesity have been the subject of numerous studies
[40][41][42][43][44][45][46][47][86,87,88,89,90,91,92,93]. The widely varying results of some studies suggest that the Pro12Ala polymorphism modulates body weight
[34][40][48][49][80,86,94,95], but that its effects are modified by environmental factors such as differences in dietary habits or levels of physical activity
[50][51][52][96,97,98].
Generally, carriers of the
PPARG 12Ala allele have significantly lower BMI values and lower fasting insulin levels with concomitant higher insulin sensitivity, as well as lower fasting plasma glucose, higher high-density lipoprotein (HDL) cholesterol, and lower total triglyceride levels
[34][53][54][80,99,100]. It has been suggested that the lower transcriptional activity of the alanine variant of the PPARγ2 receptor, which is associated with a functional repression of the transcription of genes stimulating adipogenesis, is responsible for the lower BMI values observed in 12Ala homozygotes. The slower accumulation of adipose tissue characteristic of carriers of the 12Ala allele results in better tissue response to insulin and improved insulin sensitivity
[34][80]. This hypothesis seems to be confirmed by the significantly increased activity of PPARγ2 receptors in obese individuals
[55][101]. High PPARγ2 receptor activity likely shifts the balance of metabolic changes toward fat storage and BMI increase, which may be considered a significant risk factor for the potential development of obesity in carriers of the Pro12 allele. On the other hand, the decreased activity of PPARγ2 proteins reduces the risk of developing insulin resistance, and the risk of developing type 2 diabetes in carriers of the 12Ala allele
[53][54][99,100]. This assumption has been confirmed in studies of people with type 2 diabetes, among whom people with the 12Ala allele are much less common
[34][56][80,102].
However, differential effects of the
PPARG 12Ala allele on BMI were found in overweight/obese and lean individuals. Normal body weight participants who were 12Ala homozygotes had lower BMIs, in both the study of Danish subjects
[40][86] and the previous study of Finnish non-obese subjects
[34][80]. On the other hand, 12Ala homozygotes had higher BMI values compared to Pro12 allele carriers in obese Danish participants
[40][86], which was confirmed in the American study of Caucasians with obesity, where 12Ala allele carriers had higher BMIs compared to Pro12 homozygotes
[48][94]. Moreover, a meta-analysis with 40 data sets from 30 independent studies with a total of 19136 subjects showed that in obese individuals, carriers of the Ala12 allele had higher BMI values than homozygotes for the Pro12 allele, whereas this effect was not observed in lean individuals
[49][95].
It should be also noted, however, that the biological effects of Pro12Ala variants are likely to be complex and highly dependent on the interaction between diet and gene, as well as the particular ligand involved. It is worth noting that PPARγ is a ligand-activated transcription factor that is known to be activated by dietary fatty acids. Dietary fats are important modulators of
PPARG gene transcription, leading to changes in this gene expression in response to diet. The effect of fasting was observed in mice, where it was associated with the downregulation of
Pparg expression, whereas a high-fat diet increased
Pparg expression in adipose tissue
[57][103]. Consistent with these results, a low-energy diet was shown to downregulate PPARγ mRNA expression in adipocytes from obese humans
[55][101].
All of the above data suggest that PPARG is a strong candidate gene that predisposes to obesity and that acts mainly by regulating adiposity, but also by influencing food intake and appetite control via modulating insulin-dependent signaling pathways. Moreover, the variation of the PPARG gene may play an important role in influencing the response to diet and the controls of eating behaviors. The PPARG Pro12Ala genotype is associated with susceptibility to obesity; however, the observed effects of its presence in an individual’s genotype are highly dependent on that individual’s lifestyle.
2. The PPARD Gene
Post-exercise body mass changes are controlled by a large number of genes with mild to moderate individual effects. Gene networks that are associated with BMI-related sub-phenotypes revealed nearly 100 genes that are associated with BMI phenotypic variability
[58][104]. Within the family of PPAR proteins, in addition to the previously presented PPARγ protein, another protein—peroxisome proliferator-activated receptor delta (PPARδ), encoded by the
PPARD gene—has been described as being important in the context of its role in the effectiveness of reduction diets and post-exercise weight loss
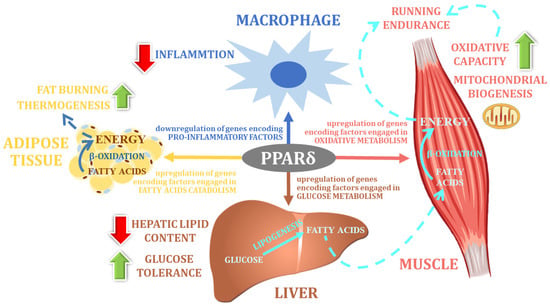
[59][45]. PPARδ, like other members of the PPAR family, is a transcription factor that regulates the expression of numerous genes encoding factors that are involved in the biochemical changes of most nutrients. Therefore, the PPARδ protein is considered to be one of the most important regulatory factors coordinating various metabolic pathways at the cellular level. Detailed studies show that this factor is particularly important for fatty acid catabolism and energy homeostasis (
Figure 23)
[60][61][62][105,106,107]. Indeed, in adipose tissue, the activation effect of PPARδ seems to be opposite to that of PPARγ: PPARδ stimulates fat burning, whereas PPARγ is mainly responsible for controlling fat storage
[59][45].
The
PPARD gene is located on human chromosome 6 at position 6p21.31
[62][63][107,108]. In contrast to
PPARA (encoding PPARα) and
PPARG (encoding PPARγ), the
PPARD gene in humans is expressed in many tissues almost throughout the body
[64][65][109,110]. A particularly high level of expression of the
PPARD gene is found in tissues where intense lipid metabolism occurs: adipose tissue, heart, and skeletal muscle. In these tissues, high levels of PPARδ protein are observed, which are necessary to stimulate the expression of genes encoding factors involved in the processes of active fatty acid uptake by cells and the β-oxidation of fatty acids
[66][111].
The activation of PPARδ factor correlates with a decrease in the plasma triglyceride levels, resulting in their lower availability and slower storage in adipose tissue
[67][112]. This suggests that proper PPARδ protein activity may be associated with a reduced susceptibility to obesity. This suggestion seems to be confirmed by studies with transgenic animals. The overexpression of the mouse
Ppard gene in adipose tissue resulted in lean mice that did not develop obesity, even when fed a high-fat diet
[68][113]. Detailed analyses in these animals showed that the Pparδ protein increased the expression of genes encoding factors involved in catabolic changes in fatty acids and adaptive thermogenesis. On the other hand, mice with a completely blocked
Ppard gene showed a marked tendency toward obesity
[68][113]. A further study with synthetic ligands activating the Pparδ protein in mice treated with a high-fat diet showed that the effect of such specific Pparδ agonists leads to the inhibition of body weight gain, but without effects on the appetite of the experimental animals and the amount of food consumed
[69][114]. This suggests the possibility of using such synthetic PPARδ agonists as potential therapeutic agents for the treatment and/or prevention of obesity—however, due to the highly pleiotropic effect of PPARδ activation
[60][70][105,115], their potential therapeutic use in humans still requires much research.
A number of single nucleotide polymorphisms (SNPs) have been described in the human
PPARD gene, and they have been analyzed with respect to their potential significance for various human traits
[71][116]. Such phenotypic modulation, due to the presence of specific polymorphic variants of the
PPARD gene, has been described in numerous scientific publications
[72][73][74][75][76][77][78][79][80][81][82][117,118,119,120,121,122,123,124,125,126,127].
The most commonly studied SNP in the context of the efficacy of dietary interventions or weight changes after exercise is rs2016520, which is located outside the region of the
PPARD gene encoding the PPARδ protein, in the regulatory region of exon 4, approximately 87 base pairs upstream of the translation start site. An in vitro study indicated that the presence of the C allele at the rs2016520 site is associated with a higher transcriptional activity of the
PPARD gene compared to the T allele, as confirmed through electrophoretic mobility shift assays
[83][128]. Given the role of the PPARδ protein in lipid metabolism, it can be assumed that the increased transcriptional activity of the
PPARD gene leading to higher levels of PPARδ protein will have an impact on the levels of lipid fractions observed in blood serum in carriers of different sequence variants of the
PPARD gene. Indeed, there are reports indicating that carriers of the rs2016520 C or T allele exhibit subtle differences in plasma LDL and HDL cholesterol, and in apoB and triglyceride concentrations
[83][84][85][86][128,129,130,131]. It has also been shown that the presence of additional polymorphic variants described at other sites in the
PPARD gene (rs6902123 in intron 2, rs2076167 in exon 7, and rs1053049 in exon 9) correlates with insulin-dependent glucose uptake by cells, and this effect appears to be exclusive to skeletal muscle, as no such correlation was observed for adipose tissue
[87][132].
Figure 23. The main physiological function of PPARδ (own elaboration, after: Reilly and Lee 2008 [88]). The main physiological function of PPARδ (own elaboration, after: Reilly and Lee 2008 [133]).
In summary, in the case of several polymorphic variants that have been described and tested in the PPARD gene, their modulatory effects on the efficacy of dietary and exercise interventions appears to be reasonably well documented. However, it is extremely difficult to specify a clear positive system of allelic variants in the PPARD gene that would provide its carrier with the greatest efficacy of a reduction diet, or the chance of the greatest weight loss through increased physical activity. At this point, it must be emphasized that the functional significance of most of the polymorphic points mentioned above (with the exception of rs2016520) is currently unknown. Therefore, to better understand the interrelationships between the individual components of the haplotype systems constructed for the PPARD gene, more detailed molecular studies are required to explain the functional significance of each of the polymorphic variants in the PPARD gene described so far.
3. The PPARA Gene
Peroxisome proliferator-activated receptor alpha (PPARα) is the last member of the PPAR family of proteins that undoubtedly plays a crucial role in the systemic regulation of metabolism. The PPARα protein is mainly expressed in liver, but also in brown adipose tissue, kidney, and cardiac and skeletal muscle
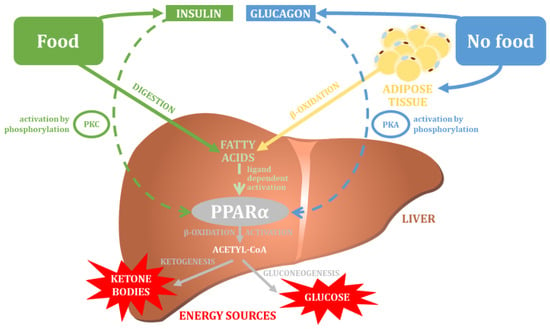
[89][141]. The main physiological function of PPARα is the regulation of fatty acid transport and their β-oxidation, which is an important element in maintaining the balance of lipid metabolism in the human body (
Figure 34)
[90][91][43,142]. More specifically, the active PPARα protein inhibits lipid accumulation by decreasing the rate of glycolysis and increasing glycogen synthesis and the aerobic metabolism of fatty acids
[92][143].
Detailed analysis shows that PPARα protein stimulates the expression of the
APOA5 gene, encoding apolipoprotein A5 (Apo-AV), which lowers blood triglyceride levels
[93][144]. The activation of PPARα decreases the plasma levels of triglyceride-rich lipoproteins primarily by stimulating expression of the
LPL gene, which encodes a lipoprotein lipase that hydrolyzes lipoprotein triglycerides
[94][145]. PPARα also causes an increase in high-density lipoprotein cholesterol (HDL-C), with a concomitant decrease in low-density lipoprotein cholesterol (LDL-C)
[95][146]. The PPARα factor mediates the increase in HDL-C levels by stimulating the expression of genes encoding apolipoproteins Apo-AI and Apo-AII
[96][147]. Furthermore, the activation of PPARα in intestinal cells decreases cholesterol esterification, inhibits chylomicron production, and increases HDL synthesis by enterocytes
[97][148].
The effect of PPARα varies depending on the nutritional state of the organism. During fasting, the PPARα protein stimulates the expression of genes encoding fatty acid transport proteins, thereby facilitating the uptake by the liver cells of free fatty acids released from adipose tissue as a result of lipolysis processes taking place there
[98][99][149,150]. PPARα activity has also been shown to promote fatty acid oxidation in the mitochondria, and peroxisomes in liver cells
[90][98][43,149]. During long periods of fasting, PPARα stimulates the formation of ketone bodies, which are used as an emergency power source in tissues outside the liver. The PPARα protein shows a slightly different effect under conditions of adequate nutrition: it is then a factor that promotes de novo lipogenesis, which involves the transfer of precursors for fatty acid synthesis (acetyl-CoA) from mitochondria to the cytosol, and aims to obtain the energy (in the form of liver triglycerides) needed to survive periods of starvation
[98][149]. Thus, the nutritional state of the organism determines the activity of the PPARα factor, which in a well-nourished organism has an activity that is related to the coordination of lipogenesis processes, while in conditions of starvation it acts as a factor that promotes the uptake of fatty acids and their oxygen conversion. It is worth mentioning that nutritional status also affects PPARα protein activity by controlling PPARα-phosphorylating kinases: during full nutrient intake, PPARα activity is stimulated by insulin-activated MAPK (mitogen-activated protein kinases) and glucose-activated PKC (protein kinase C), whereas during fasting periods, PPARα is activated by glucagon-activated PKA (protein kinase A) and AMPK (5′AMP-activated protein kinase)
[99][150].
Figure 34. The main physiological function of PPARα (own elaboration, after: Pawlak et al. 2015
[99][150]). PKC—protein kinase C; PKA—protein kinase A.
Considering the importance of PPARα in the context of the effect of this transcription factor on the efficacy of applied reduction diets and the effectiveness of weight reduction after exercise, it should be noted that the activation of PPARα stimulates weight reduction in rodents
[100][151], but that this effect has not been clearly confirmed in humans—keeping in mind that studies that do not confirm this phenomenon have so far been conducted mainly in patients with type 2 diabetes
[101][102][152,153], and that there are no such studies in healthy humans. On the other hand, there is also a study suggesting that the activation of PPARα in the central nervous system can lead to increased food intake
[103][154], but there is also evidence of the opposite effects, where the activation of PPARα-dependent pathways has been shown to induce satiety and reduce appetite
[104][155].
The
PPARA gene is located on human chromosome 22 at position 22q13.31
[105][156]. Many SNP points have been identified within the human
PPARA gene
[106] (Table 1) [157]; one of the most frequently described being the C-to-G transversion, the consequence of which is an amino acid substitution at position 162 (Leu162Val; rs1800206) of human PPARα
[107][108][158,159]. This amino acid change has real functional significance because it occurs in the region of the
PPARA gene that encodes the DNA-binding domain, namely, the second zinc finger. The conversion of valine to leucine occurs directly adjacent to the cysteine that is required to coordinate the Zn
2+ atom of the second zinc finger, i.e., upstream of the region that determines the specificity and polarity of PPARα binding to DNA in the target genes
[109][160]. There are evidences indicating that the variant of the PPARα protein with valine at position 162 (allele 162Val) is a more active transcription factor than the form with Leu162
[110][161]. However, it should be emphasized that this functional diversity depends on the concentration of PPARα ligands, which may be, for example, free fatty acids. In the absence of or at low concentrations of ligands, the activity of the 162Val variant is almost half that of the leucine form. In contrast, at high ligand concentrations, the activity of the valine form increases significantly and may even exceed that of the Leu162 variant
[110][161].
There is ample evidence that the Leu162Val polymorphism plays a role in shaping lipid metabolism. Several studies suggest that the presence of the 162Val allele is positively correlated with elevated total cholesterol, LDL-C, triglyceride, and Apo-B levels, which translates into an increased risk of hyperlipidemia and obesity
[107][111][112][113][158,162,163,164]. On the other hand, there are reports that do not confirm the association between Leu162Val polymorphism and body mass index, body fat composition, liver fat content, or more generally with obesity
[114][115][165,166].
As with
PPARG gene diversity, the functional significance of the
PPARA polymorphic variants may be altered by various environmental factors, including diet. An intervention study suggested that carriers of the 162Val allele have higher levels of triglycerides and Apo-CIII, but only if their diet was low in polyunsaturated fatty acids. The opposite effect was observed when the diet was enriched in polyunsaturated fatty acids
[116][167]. Similar effects were found in another study, which showed that carriers of the 162Val allele had a lower total cholesterol and Apo-AI levels in diets rich in polyunsaturated fatty acids
[117][168].
Another polymorphic point in the
PPARA gene that is of functional significance is Val227Ala (rs1800234), which appears to be present primarily in individuals of oriental descent
[118][119][169,170]. In this case, the Val/Ala amino acid substitution, which is a consequence of the nucleotide change in the
PPARA gene, occurs in the so-called hinge region, which is located between the DNA-binding domain (DBD) and the ligand-binding domain (LBD). The dimerization domain of the protein is located in this region, and therefore, the conversion of valine to alanine at position 227 is thought to be of functional importance: the 227Ala variant is characterized by a lower transactivation activity than the Val227 form in the presence of PPARα-specific ligands
[120][171]. An in vitro study suggested that the 227Ala allele is associated with a slower transcription of the cytochrome P-450 4A6 gene and the mitochondrial 3HMG-CoA synthase gene
[120][171]. In addition, the alanine variant is positively correlated with reduced levels of cholesterol, LDL-C, and triglycerides
[118][119][169,170]—thus, the 227Ala allele is considered as being important for lipid lowering and protection against the development of steatosis
[121][172]. The Val227 variant, on the other hand, is associated with higher cholesterol levels—an effect that occurs primarily in individuals who do not drink alcohol. In alcoholic drinkers, the 227Ala allele appears to be associated with increased gamma-glutamyl transpeptidase activity
[122][173].
Among noncoding SNPs, the most commonly studied is the point in intron 7 of the
PPARA gene, which is a G-to-C transversion (rs4253778)
[123][124][174,175]. Although this is a non-functional polymorphism (causing no changes in the amino acid sequence of the encoded protein), there are reports indicating that the presence of specific alleles at this SNP point is associated with changes in the expression of the
PPARA gene itself, such that the C allele correlates with a lower expression of the
PPARA gene in its carriers
[125][176], which in turn may lead to a weaker stimulation of the expression of genes controlled by PPARα
[126][177]. CC homozygotes have significantly higher total and LDL cholesterol levels
[127][178], and the C allele showed a positive association with dyslipidemia
[128][179]. On the other hand, carriers of the C allele were found to have significantly lower TG and VLDL levels and higher HDL-C levels in populations of different ethnic origins
[129][180].
Another noncoding SNP worth mentioning is the T in A nucleotide change in the intron of the
PPARA gene, designated rs4253747. This change appears to be of particular interest from the point of view of the efficacy of the reduction diets used, as it has been shown that carriers of the A allele are characterized by a much stronger appetite suppression response that occurs under certain conditions, described as high-altitude appetite loss, which is a common symptom of acute mountain sickness
[130][181]. Exposure to high altitude is thought to stimulate
PPARA gene expression
[131][132][182,183]—the involvement of rs4253747 variants in this process is not yet known, but it is expected that they may be important for binding transcription factors within the
PPARA gene promoter or enhancers, such that they control the expression of the
PPARA gene itself
[130][181].
There are also polymorphic points in the
PPARA 3′UTR region (the gene region encoding the so-called untranslated region), such as rs6008259 and rs3892755, for which clear interactions between the presence of specific variants, diet, and metabolic parameters have been demonstrated. With respect to the SNP rs6008259, it has been shown that higher levels of total cholesterol and LDL-C are observed in carriers of the G allele at a high daily intake of linolenic acid, compared with carriers of the genotype AA. Similarly, carriers of the C allele described in item rs3892755 with a high daily intake of eicosapentaenoic acid and docosahexaenoic acid are characterized by high levels of total cholesterol and LDL-C, compared with carriers of the TT genotype
[106][133][157,184].
The PPARα protein is undoubtedly an important metabolic regulator. As mentioned earlier, the diversity of the PPARA gene affects the content of different lipid fractions and related particles observed in the blood plasma of carriers of different polymorphic variants. The sequence variants described at the different polymorphic sites of the PPARA gene can therefore be considered as promising markers that could potentially be used to predict the effects of appropriate nutritional interventions.
4. The PPARGC1A Gene
Coordinated metabolic regulation is undoubtedly one of the key factors in maintaining a healthy body weight. PPARs are an important element of this regulation, but they are not the only important factors influencing BMI and related values. Specific proteins, known as coactivators, are required for the activity of PPARs and many other transcription factors. One such coactivator is peroxisome proliferator-activated receptor gamma coactivator-1-alpha (PGC-1α), which in humans is encoded by the
PPARGC1A gene
[134][185].
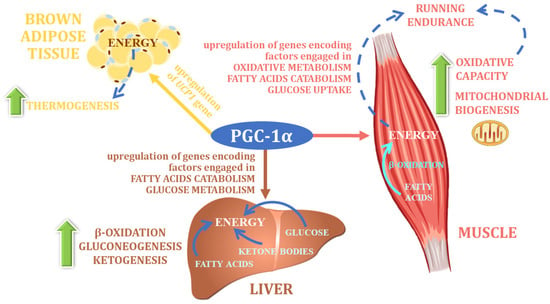
PGC-1α is considered to be the central regulator of energy metabolism (
Figure 45). It was originally described as a cold-inducible coactivator of nuclear receptors associated with adaptive thermogenesis, as the first study on this factor indicated that Pgc-1α mRNA expression is dramatically increased upon cold exposure of mice, in both brown fat and skeletal muscle, tissues that represent the main centers of thermogenesis in mammals
[135][186]. PGC-1α has been shown to control oxidative metabolism in many cell types
[136][187]. The expression of the
Ppargc1a gene encoding this factor in mice is induced under the influence of physical activity in muscle, where it stimulates mitochondrial biogenesis
[137][188], angiogenesis
[138][189], and the transformation of muscle fibers toward their greater oxygen capacity
[139][190]. Changes in the metabolic profile observed in the muscles of physically active individuals due to increased expression of the
PPARGC1A gene include not only increased capillarization, muscle fiber remodeling, or mitochondrial biogenesis, but also the pleiotropic control of many pathways of energy substrate conversion, such as fatty acid oxidation, tricarboxylic acid cycle (TCA), and glucose transport pathways, as well as a significant increase in the level and rate of oxidative phosphorylation
[136][187].
Figure 45. The main physiological function of PGC-1α (own elaboration, after: Cheng et a. 2018
[140][16]). UCP1—uncoupling protein 1.
Contrary to its name, PGC-1α is not only a PPARγ coactivator, but also other transcription factors that are involved in the control of the expression of numerous key genes, not only in terms of mitochondrial biogenesis or angiogenesis, but also in terms of adipose tissue metabolism and the potential risk of developing obesity. In the late 1990s, Puigserver et al. found that obesity is the result of an imbalance between energy production (derived from calories consumed with food) and energy expenditure
[135][186]. Total energy expenditure includes: basal metabolic rate, dietary energy expenditure, the energy cost of physical activity, and what is known as adaptive (non-shivering) thermogenesis
[141][191]. The latter process refers to energy that is expended in response to changing environmental conditions, particularly when the body is exposed to cold or excessive caloric intake (diet-induced thermogenesis). Adaptive thermogenesis is an important component of energy homeostasis, and it is considered as being part of the metabolic defense against obesity
[135][186].
Long-standing research has shown that PGC-1α is the master regulator of uncoupling protein 1 (UCP1) expression, which activates thermogenic programs in brown adipose tissue and elsewhere
[142][192]. PGC-1α not only directly controls the expression of UCP-1 protein, but also stimulates the expression of several products secreted by muscle during exercise. One of these products is type I membrane protein, a precursor of irisin, which strongly stimulates the expression of UCP-1 and the functional conversion of white adipose tissue cells into brown fat cells
[143][193]. These processes lead to a significant increase in total energy expenditure, mainly due to an increase in the component related to adaptive thermogenesis, which translates into a lower tendency towards increased weight gain, and a lower risk of obesity. This assumption seems to be confirmed by a study with transgenic mice, in which the muscular expression of the Pgc-1α protein was increased—a prolonged lifespan and resistance to age-related obesity and diabetes were observed
[144][194]. Therefore, it can be assumed that carriers of the allelic variants of the
PPARGC1A gene that favor a higher expression of this gene and an increased level of PGC-1α protein are characterized by a higher energy expenditure that is associated with thermogenic changes, and consequently, by a lower probability of weight gain and the development of obesity. It is worth mentioning that the thermogenic function of PGC-1α is similar to the effect of PPAR-δ mentioned above.
The
PPARGC1A gene was fully described in the late 1990s
[134][135][185,186]. Since then, numerous polymorphic variants of this gene have been described in humans, including those of functional significance. The most commonly described polymorphic point in the
PPARGC1A gene is the G-to-A transition at position +1564 in exon 8 (rs8192678), which is known as the Gly482Ser substitution
[145][195]. Studies with different groups of participants suggest that the 482Ser allele is associated with a reduction in
PPARGC1A gene expression, resulting in lower mRNA levels in carriers of this variant
[146][147][148][196,197,198]. In vitro experiments suggest that the amino acid change at position 482 at the molecular level decreases the ability of PGC-1α to bind to the MEF2C protein, which is a transcription factor that is involved in controlling the expression of many genes
[149][199]. It has been shown that the efficiency of the interaction between coactivator and transcription factor is significantly reduced when serine is present at position 482 (in the domain responsible for the interaction between PGC-1α and MEF2C) instead of glycine
[149][150][199,200]. In mice, the activation of Mef2c by Pgc-1α protein has been shown to play an autoregulatory role, as the Pgc-1α/Mef2c complex enhances expression of the gene encoding mouse Pgc-1α protein
[151][201].